The Influence of Sodium Chloride on the Performance of Gammarus Amphipods and the Community Composition of Microbes Associated with Leaf Detritus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Isabel Cristina Santos Silva De Faria Ramos Comunidade Bacteriana
Universidade de Aveiro Departamento de Biologia 2009 Isabel Cristina Santos Comunidade bacteriana cultivável da microcamada Silva de Faria Ramos superficial estuarina Culturable bacterial community of the estuarine surface microlayer Universidade de Aveiro Departamento de Biologia 2009 Isabel Cristina Santos Comunidade bacteriana cultivável da microcamada Silva de Faria Ramos superficial estuarina Culturable bacterial community of the estuarine surface microlayer dissertação apresentada à Universidade de Aveiro para cumprimento dos requisitos necessários à obtenção do grau de Mestre em Microbiologia, realizada sob a orientação científica da Prof. Dra. Isabel Henriques, Professora Auxiliar Convidada do Departamento de Biologia da Universidade de Aveiro. Dedico este trabalho à minha família por todo o apoio e compreensão. o júri presidente Prof. Doutora Sónia Alexandra Leite Velho Mendo Barroso professora auxiliar do Departamento de Biologia da Universidade de Aveiro Prof. Doutor Fernando Manuel dos Santos Tavares professor auxiliar do Departamento de Botânica, Faculdade de Ciências da Universidade do Porto Prof. Doutora Isabel da Silva Henriques professora auxiliar convidada do Departamento de Biologia da Universidade de Aveiro agradecimentos A primeira pessoa a quem quero agradecer é ao Professor António Correia pela oportunidade de desenvolver este trabalho no seu laboratório e pelo exemplo de sacrifício e constante optimismo com que temos que enfrentar a vida! Quero agradecer à minha orientadora, Doutora Isabel Henriques, com quem mantive um relação cordial e leal durante todo o trabalho, por tudo o que me ensinou…que foi muito mais além do que conhecimento científico. Aprendi a enfrentar as agruras do trabalho com perseverança e entusiasmo. A todos os meus colegas de laboratório com quem convivi e partilhei todas as minhas alegrias e frustrações. -

Genomics 98 (2011) 370–375
Genomics 98 (2011) 370–375 Contents lists available at ScienceDirect Genomics journal homepage: www.elsevier.com/locate/ygeno Whole-genome comparison clarifies close phylogenetic relationships between the phyla Dictyoglomi and Thermotogae Hiromi Nishida a,⁎, Teruhiko Beppu b, Kenji Ueda b a Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan b Life Science Research Center, College of Bioresource Sciences, Nihon University, Fujisawa, Japan article info abstract Article history: The anaerobic thermophilic bacterial genus Dictyoglomus is characterized by the ability to produce useful Received 2 June 2011 enzymes such as amylase, mannanase, and xylanase. Despite the significance, the phylogenetic position of Accepted 1 August 2011 Dictyoglomus has not yet been clarified, since it exhibits ambiguous phylogenetic positions in a single gene Available online 7 August 2011 sequence comparison-based analysis. The number of substitutions at the diverging point of Dictyoglomus is insufficient to show the relationships in a single gene comparison-based analysis. Hence, we studied its Keywords: evolutionary trait based on whole-genome comparison. Both gene content and orthologous protein sequence Whole-genome comparison Dictyoglomus comparisons indicated that Dictyoglomus is most closely related to the phylum Thermotogae and it forms a Bacterial systematics monophyletic group with Coprothermobacter proteolyticus (a constituent of the phylum Firmicutes) and Coprothermobacter proteolyticus Thermotogae. Our findings indicate that C. proteolyticus does not belong to the phylum Firmicutes and that the Thermotogae phylum Dictyoglomi is not closely related to either the phylum Firmicutes or Synergistetes but to the phylum Thermotogae. © 2011 Elsevier Inc. -

New Zealand's Genetic Diversity
1.13 NEW ZEALAND’S GENETIC DIVERSITY NEW ZEALAND’S GENETIC DIVERSITY Dennis P. Gordon National Institute of Water and Atmospheric Research, Private Bag 14901, Kilbirnie, Wellington 6022, New Zealand ABSTRACT: The known genetic diversity represented by the New Zealand biota is reviewed and summarised, largely based on a recently published New Zealand inventory of biodiversity. All kingdoms and eukaryote phyla are covered, updated to refl ect the latest phylogenetic view of Eukaryota. The total known biota comprises a nominal 57 406 species (c. 48 640 described). Subtraction of the 4889 naturalised-alien species gives a biota of 52 517 native species. A minimum (the status of a number of the unnamed species is uncertain) of 27 380 (52%) of these species are endemic (cf. 26% for Fungi, 38% for all marine species, 46% for marine Animalia, 68% for all Animalia, 78% for vascular plants and 91% for terrestrial Animalia). In passing, examples are given both of the roles of the major taxa in providing ecosystem services and of the use of genetic resources in the New Zealand economy. Key words: Animalia, Chromista, freshwater, Fungi, genetic diversity, marine, New Zealand, Prokaryota, Protozoa, terrestrial. INTRODUCTION Article 10b of the CBD calls for signatories to ‘Adopt The original brief for this chapter was to review New Zealand’s measures relating to the use of biological resources [i.e. genetic genetic resources. The OECD defi nition of genetic resources resources] to avoid or minimize adverse impacts on biological is ‘genetic material of plants, animals or micro-organisms of diversity [e.g. genetic diversity]’ (my parentheses). -
The RDP-II Backbone Tree for Release 8.0. the Tree Was Inferred from a Distance Matrix Generated in PAUP* with the Weighbor (Weighted Neighbor Joining) Algorithm
Methylococcus capsulatus ACM 1292 (T) Oceanospirillum linum ATCC 11336 (T) Halomonas halodenitrificans ATCC 13511 (T) Legionella lytica PCM 2298 (T) Francisella tularensis subsp. tularensis ATCC 6223 (T) Coxiella burnetii Q177 Moraxella catarrhalis ATCC 25238 (T) Pseudomonas fluorescens IAM 12022 (T) Piscirickettsia salmonis LF-89 (T) Thiothrix nivea DSM 5205 (T) Allochromatium minutissimum DSM 1376 (T) Alteromonas macleodii IAM 12920 (T) Aeromonas salmonicida subsp. smithia CCM 4103 (T) Pasteurella multocida NCTC 10322 (T) Enterobacter nimipressuralis LMG 10245 (T) Vibrio vulnificus ATCC 27562 (T) Ectothiorhodospira mobilis DSM237 (T) Xanthomonas campestris LMG 568 (T) Cardiobacterium hominis ATCC 15826 (T) Methylophilus methylotrophus ATCC 53528 (T) Rhodocyclus tenuis DSM 109 (T) Hydrogenophilus thermoluteolus TH-1 (T) Neisseria gonorrhoeae NCTC 8375 (T) Comamonas testosteroni ATCC 11996 (T) Nitrosospira multiformis ATCC 25196 (T) Spirillum volutans ATCC 19554 (T) Burkholderia glathei LMG 14190 (T) Alcaligenes defragrans DSM 12141 (T) Oxalobacter formigenes ATCC 35274 (T) Acetobacter oboediens DSM 11826 (T) clone CS93 PROTEOBACTERIA Caedibacter caryophilus 221 (T) Rhodobacter sphaeroides ATCC 17023 (T) Rickettsia rickettsii ATCC VR-891 (T) Ehrlichia risticii ATCC VR-986 (T) Sphingomonas paucimobilis GIFU 2395 (T) Caulobacter fusiformis ATCC 15257 (T) Rhodospirillum rubrum ATCC 11170 (T) Brucella melitensis ATCC 23459 (T) Rhizobium tropici IFO 15247 (T) Bartonella vinsonii subsp. vinsonii ATCC VR-152 (T) Phyllobacterium myrsinacearum -

Leeuwenhoekiella Palythoae Sp. Nov., a New Member of the Family Flavobacteriaceae
International Journal of Systematic and Evolutionary Microbiology (2009), 59, 3074–3077 DOI 10.1099/ijs.0.010371-0 Leeuwenhoekiella palythoae sp. nov., a new member of the family Flavobacteriaceae Olga I. Nedashkovskaya,1 Marc Vancanneyt,2 Seung Bum Kim,3 Natalia V. Zhukova,4 Ji Hye Han3 and Valery V. Mikhailov1 Correspondence 1Pacific Institute of Bioorganic Chemistry of the Far-Eastern Branch of the Russian Academy of Olga I. Nedashkovskaya Sciences, Pr. 100 Let Vladivostoku 159, 690022 Vladivostok, Russia [email protected] 2BCCM/LMG Bacteria Collection, and Laboratory of Microbiology, Ghent University, Ledeganckstraat 35, B-9000 Ghent, Belgium 3Department of Microbiology, School of Bioscience and Biotechnology, Chungnam National University, 220 Gung-dong, Yuseong, Daejeon 305-764, Republic of Korea 4Institute of Marine Biology of the Far-Eastern Branch of the Russian Academy of Sciences, Pal’chevskogo St. 17, 690032, Vladivostok, Russia The taxonomic status of a novel, heterotrophic, strictly aerobic, gliding and yellow–orange- pigmented bacterium (strain KMM 6264T), associated with the coral Palythoa, was determined. The 16S rRNA gene sequence analysis indicated that strain KMM 6264T clustered with the recognized species of the genus Leeuwenhoekiella of the family Flavobacteriaceae with 96.4– 98.2 % sequence similarity. DNA–DNA reassociation levels between the isolate and the type strains of Leeuwenhoekiella species were 15–22 %. The DNA G+C content was 41.2 mol%. The phylogenetic evidence and the results of genomic and phenotypic analyses showed that the isolate should be classified as a member of a novel species of the genus Leeuwenhoekiella, for which the name Leeuwenhoekiella palythoae sp. nov. -

Comparative Proteomic Profiling of Newly Acquired, Virulent And
www.nature.com/scientificreports OPEN Comparative proteomic profling of newly acquired, virulent and attenuated Neoparamoeba perurans proteins associated with amoebic gill disease Kerrie Ní Dhufaigh1*, Eugene Dillon2, Natasha Botwright3, Anita Talbot1, Ian O’Connor1, Eugene MacCarthy1 & Orla Slattery4 The causative agent of amoebic gill disease, Neoparamoeba perurans is reported to lose virulence during prolonged in vitro maintenance. In this study, the impact of prolonged culture on N. perurans virulence and its proteome was investigated. Two isolates, attenuated and virulent, had their virulence assessed in an experimental trial using Atlantic salmon smolts and their bacterial community composition was evaluated by 16S rRNA Illumina MiSeq sequencing. Soluble proteins were isolated from three isolates: a newly acquired, virulent and attenuated N. perurans culture. Proteins were analysed using two-dimensional electrophoresis coupled with liquid chromatography tandem mass spectrometry (LC–MS/MS). The challenge trial using naïve smolts confrmed a loss in virulence in the attenuated N. perurans culture. A greater diversity of bacterial communities was found in the microbiome of the virulent isolate in contrast to a reduction in microbial community richness in the attenuated microbiome. A collated proteome database of N. perurans, Amoebozoa and four bacterial genera resulted in 24 proteins diferentially expressed between the three cultures. The present LC–MS/ MS results indicate protein synthesis, oxidative stress and immunomodulation are upregulated in a newly acquired N. perurans culture and future studies may exploit these protein identifcations for therapeutic purposes in infected farmed fsh. Neoparamoeba perurans is an ectoparasitic protozoan responsible for the hyperplastic gill infection of marine cultured fnfsh referred to as amoebic gill disease (AGD)1. -

Metatranscriptomic Analysis of Community Structure And
School of Environmental Sciences Metatranscriptomic analysis of community structure and metabolism of the rhizosphere microbiome by Thomas Richard Turner Submitted in partial fulfilment of the requirement for the degree of Doctor of Philosophy, September 2013 This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that use of any information derived there from must be in accordance with current UK Copyright Law. In addition, any quotation or extract must include full attribution. i Declaration I declare that this is an account of my own research and has not been submitted for a degree at any other university. The use of material from other sources has been properly and fully acknowledged, where appropriate. Thomas Richard Turner ii Acknowledgements I would like to thank my supervisors, Phil Poole and Alastair Grant, for their continued support and guidance over the past four years. I’m grateful to all members of my lab, both past and present, for advice and friendship. Graham Hood, I don’t know how we put up with each other, but I don’t think I could have done this without you. Cheers Salt! KK, thank you for all your help in the lab, and for Uma’s biryanis! Andrzej Tkatcz, thanks for the useful discussions about our projects. Alison East, thank you for all your support, particularly ensuring Graham and I did not kill each other. I’m grateful to Allan Downie and Colin Murrell for advice. For sequencing support, I’d like to thank TGAC, particularly Darren Heavens, Sophie Janacek, Kirsten McKlay and Melanie Febrer, as well as John Walshaw, Mark Alston and David Swarbreck for bioinformatic support. -

Table S4. Phylogenetic Distribution of Bacterial and Archaea Genomes in Groups A, B, C, D, and X
Table S4. Phylogenetic distribution of bacterial and archaea genomes in groups A, B, C, D, and X. Group A a: Total number of genomes in the taxon b: Number of group A genomes in the taxon c: Percentage of group A genomes in the taxon a b c cellular organisms 5007 2974 59.4 |__ Bacteria 4769 2935 61.5 | |__ Proteobacteria 1854 1570 84.7 | | |__ Gammaproteobacteria 711 631 88.7 | | | |__ Enterobacterales 112 97 86.6 | | | | |__ Enterobacteriaceae 41 32 78.0 | | | | | |__ unclassified Enterobacteriaceae 13 7 53.8 | | | | |__ Erwiniaceae 30 28 93.3 | | | | | |__ Erwinia 10 10 100.0 | | | | | |__ Buchnera 8 8 100.0 | | | | | | |__ Buchnera aphidicola 8 8 100.0 | | | | | |__ Pantoea 8 8 100.0 | | | | |__ Yersiniaceae 14 14 100.0 | | | | | |__ Serratia 8 8 100.0 | | | | |__ Morganellaceae 13 10 76.9 | | | | |__ Pectobacteriaceae 8 8 100.0 | | | |__ Alteromonadales 94 94 100.0 | | | | |__ Alteromonadaceae 34 34 100.0 | | | | | |__ Marinobacter 12 12 100.0 | | | | |__ Shewanellaceae 17 17 100.0 | | | | | |__ Shewanella 17 17 100.0 | | | | |__ Pseudoalteromonadaceae 16 16 100.0 | | | | | |__ Pseudoalteromonas 15 15 100.0 | | | | |__ Idiomarinaceae 9 9 100.0 | | | | | |__ Idiomarina 9 9 100.0 | | | | |__ Colwelliaceae 6 6 100.0 | | | |__ Pseudomonadales 81 81 100.0 | | | | |__ Moraxellaceae 41 41 100.0 | | | | | |__ Acinetobacter 25 25 100.0 | | | | | |__ Psychrobacter 8 8 100.0 | | | | | |__ Moraxella 6 6 100.0 | | | | |__ Pseudomonadaceae 40 40 100.0 | | | | | |__ Pseudomonas 38 38 100.0 | | | |__ Oceanospirillales 73 72 98.6 | | | | |__ Oceanospirillaceae -

Within-Arctic Horizontal Gene Transfer As a Driver of Convergent Evolution in Distantly Related 1 Microalgae 2 Richard G. Do
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.31.454568; this version posted August 2, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Within-Arctic horizontal gene transfer as a driver of convergent evolution in distantly related 2 microalgae 3 Richard G. Dorrell*+1,2, Alan Kuo3*, Zoltan Füssy4, Elisabeth Richardson5,6, Asaf Salamov3, Nikola 4 Zarevski,1,2,7 Nastasia J. Freyria8, Federico M. Ibarbalz1,2,9, Jerry Jenkins3,10, Juan Jose Pierella 5 Karlusich1,2, Andrei Stecca Steindorff3, Robyn E. Edgar8, Lori Handley10, Kathleen Lail3, Anna Lipzen3, 6 Vincent Lombard11, John McFarlane5, Charlotte Nef1,2, Anna M.G. Novák Vanclová1,2, Yi Peng3, Chris 7 Plott10, Marianne Potvin8, Fabio Rocha Jimenez Vieira1,2, Kerrie Barry3, Joel B. Dacks5, Colomban de 8 Vargas2,12, Bernard Henrissat11,13, Eric Pelletier2,14, Jeremy Schmutz3,10, Patrick Wincker2,14, Chris 9 Bowler1,2, Igor V. Grigoriev3,15, and Connie Lovejoy+8 10 11 1 Institut de Biologie de l'ENS (IBENS), Département de Biologie, École Normale Supérieure, CNRS, 12 INSERM, Université PSL, 75005 Paris, France 13 2CNRS Research Federation for the study of Global Ocean Systems Ecology and Evolution, 14 FR2022/Tara Oceans GOSEE, 3 rue Michel-Ange, 75016 Paris, France 15 3 US Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 16 Cyclotron Road, Berkeley, -
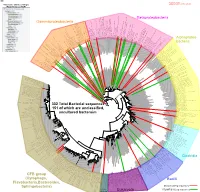
Bacteria Clostridia Bacilli Eukaryota CFB Group
AM935842.1.1361 uncultured Burkholderiales bacterium Class Betaproteobacteria AY283260.1.1552 Alcaligenes sp. PCNB−2 Class Betaproteobacteria AM934953.1.1374 uncultured Burkholderiales bacterium Class Betaproteobacteria AJ581593.1.1460 uncultured betaAM936569.1.1351 proteobacterium uncultured Class Betaproteobacteria Derxia sp. Class Betaproteobacteria AJ581621.1.1418 uncultured beta proteobacterium Class Betaproteobacteria DQ248272.1.1498 uncultured soil bacterium soil uncultured DQ248272.1.1498 DQ248235.1.1498 uncultured soil bacterium RS49 DQ248270.1.1496 uncultured soil bacterium DQ256489.1.1211 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax DQ256489.1.1211 AF523053.1.1486 uncultured Comamonadaceae bacterium Class Betaproteobacteria AY706442.1.1396 uncultured bacterium uncultured AY706442.1.1396 AJ536763.1.1422 uncultured bacterium CS000359.1.1530 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax CS000359.1.1530 AY168733.1.1411 uncultured bacterium AJ009470.1.1526 uncultured bacterium SJA−62 Class Betaproteobacteria Class SJA−62 bacterium uncultured AJ009470.1.1526 AY212561.1.1433 uncultured bacterium D16212.1.1457 Rhodoferax fermentans Class Betaproteobacteria Class fermentans Rhodoferax D16212.1.1457 AY957894.1.1546 uncultured bacterium AJ581620.1.1452 uncultured beta proteobacterium Class Betaproteobacteria RS76 AY625146.1.1498 uncultured bacterium RS65 DQ316832.1.1269 uncultured beta proteobacterium Class Betaproteobacteria DQ404909.1.1513 uncultured bacterium uncultured DQ404909.1.1513 AB021341.1.1466 bacterium rM6 AJ487020.1.1500 uncultured bacterium uncultured AJ487020.1.1500 RS7 RS86RC AF364862.1.1425 bacterium BA128 Class Betaproteobacteria AY957931.1.1529 uncultured bacterium uncultured AY957931.1.1529 CP000884.723807.725332 Delftia acidovorans SPH−1 Class Betaproteobacteria AY957923.1.1520 uncultured bacterium uncultured AY957923.1.1520 RS18 AY957918.1.1527 uncultured bacterium uncultured AY957918.1.1527 AY945883.1.1500 uncultured bacterium AF526940.1.1489 uncultured Ralstonia sp. -

United States Patent (10) Patent No.: US 7820,184 B2 Stritzker Et Al
USOO782O184B2 (12) United States Patent (10) Patent No.: US 7820,184 B2 Stritzker et al. (45) Date of Patent: Oct. 26, 2010 (54) METHODS AND COMPOSITIONS FOR 5,833,975 A 1 1/1998 Paoletti et al. ............. 424.93.2 DETECTION OF MICROORGANISMS AND 5,976,796. A 1 1/1999 Szalay et al. ................... 435/6 SirNRTREATMENT OF DISEASES AND 6,025,155 A 2/2000 Hadlaczky et al. ......... 435/69.1 6,045,802 A 4/2000 Schlom et al. ........... 424,199.1 (75) Inventors: Jochen Harald Stritzker, Kissing (DE); 6,077,697 A 6/2000 Hadlaczky et al. 435/1723 Phil Hill, West Bridgford (GB); Aladar 6,080,849 A 6/2000 Bermudes et al. .......... 536,23.7 A. Szalay, Highland, CA (US); Yong A. 6,093,700 A 7/2000 Mastrangelo et al. ......... 514,44 Yu, San Diego, CA (US) 6,099,848. A 8/2000 Frankel et al. ........... 424,246.1 6,106,826 A 8/2000 Brandt et al. .............. 424.93.2 (73) Assignee: stylus Corporation, San Diego, CA 6, 190,657 B1 2/2001 Pawelek et al. ............ 424,931 6,217,847 B1 4/2001 Contaget al. ................ 4249.1 (*) Notice: Subject to any disclaimer, the term of this 6,232,523 B1 5/2001 Tan et al. ...................... 800, 10 patent is extended or adjusted under 35 6,235,967 B1 5/2001 Tan et al. ...................... 800, 10 U.S.C. 154(b) by 362 days. 6,235,968 B1 5/2001 Tan et al. ...................... 800, 10 6,251,384 B1 6/2001 Tan et al. -

A Genomic Journey Through a Genus of Large DNA Viruses
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Virology Papers Virology, Nebraska Center for 2013 Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses Adrien Jeanniard Aix-Marseille Université David D. Dunigan University of Nebraska-Lincoln, [email protected] James Gurnon University of Nebraska-Lincoln, [email protected] Irina V. Agarkova University of Nebraska-Lincoln, [email protected] Ming Kang University of Nebraska-Lincoln, [email protected] See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/virologypub Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cell and Developmental Biology Commons, Genetics and Genomics Commons, Infectious Disease Commons, Medical Immunology Commons, Medical Pathology Commons, and the Virology Commons Jeanniard, Adrien; Dunigan, David D.; Gurnon, James; Agarkova, Irina V.; Kang, Ming; Vitek, Jason; Duncan, Garry; McClung, O William; Larsen, Megan; Claverie, Jean-Michel; Van Etten, James L.; and Blanc, Guillaume, "Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses" (2013). Virology Papers. 245. https://digitalcommons.unl.edu/virologypub/245 This Article is brought to you for free and open access by the Virology, Nebraska Center for at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Virology Papers by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Adrien Jeanniard, David D. Dunigan, James Gurnon, Irina V. Agarkova, Ming Kang, Jason Vitek, Garry Duncan, O William McClung, Megan Larsen, Jean-Michel Claverie, James L. Van Etten, and Guillaume Blanc This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ virologypub/245 Jeanniard, Dunigan, Gurnon, Agarkova, Kang, Vitek, Duncan, McClung, Larsen, Claverie, Van Etten & Blanc in BMC Genomics (2013) 14.