Db20-0503.Full.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio In
microorganisms Review The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease Spase Stojanov 1,2, Aleš Berlec 1,2 and Borut Štrukelj 1,2,* 1 Faculty of Pharmacy, University of Ljubljana, SI-1000 Ljubljana, Slovenia; [email protected] (S.S.); [email protected] (A.B.) 2 Department of Biotechnology, Jožef Stefan Institute, SI-1000 Ljubljana, Slovenia * Correspondence: borut.strukelj@ffa.uni-lj.si Received: 16 September 2020; Accepted: 31 October 2020; Published: 1 November 2020 Abstract: The two most important bacterial phyla in the gastrointestinal tract, Firmicutes and Bacteroidetes, have gained much attention in recent years. The Firmicutes/Bacteroidetes (F/B) ratio is widely accepted to have an important influence in maintaining normal intestinal homeostasis. Increased or decreased F/B ratio is regarded as dysbiosis, whereby the former is usually observed with obesity, and the latter with inflammatory bowel disease (IBD). Probiotics as live microorganisms can confer health benefits to the host when administered in adequate amounts. There is considerable evidence of their nutritional and immunosuppressive properties including reports that elucidate the association of probiotics with the F/B ratio, obesity, and IBD. Orally administered probiotics can contribute to the restoration of dysbiotic microbiota and to the prevention of obesity or IBD. However, as the effects of different probiotics on the F/B ratio differ, selecting the appropriate species or mixture is crucial. The most commonly tested probiotics for modifying the F/B ratio and treating obesity and IBD are from the genus Lactobacillus. In this paper, we review the effects of probiotics on the F/B ratio that lead to weight loss or immunosuppression. -

Genomics 98 (2011) 370–375
Genomics 98 (2011) 370–375 Contents lists available at ScienceDirect Genomics journal homepage: www.elsevier.com/locate/ygeno Whole-genome comparison clarifies close phylogenetic relationships between the phyla Dictyoglomi and Thermotogae Hiromi Nishida a,⁎, Teruhiko Beppu b, Kenji Ueda b a Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan b Life Science Research Center, College of Bioresource Sciences, Nihon University, Fujisawa, Japan article info abstract Article history: The anaerobic thermophilic bacterial genus Dictyoglomus is characterized by the ability to produce useful Received 2 June 2011 enzymes such as amylase, mannanase, and xylanase. Despite the significance, the phylogenetic position of Accepted 1 August 2011 Dictyoglomus has not yet been clarified, since it exhibits ambiguous phylogenetic positions in a single gene Available online 7 August 2011 sequence comparison-based analysis. The number of substitutions at the diverging point of Dictyoglomus is insufficient to show the relationships in a single gene comparison-based analysis. Hence, we studied its Keywords: evolutionary trait based on whole-genome comparison. Both gene content and orthologous protein sequence Whole-genome comparison Dictyoglomus comparisons indicated that Dictyoglomus is most closely related to the phylum Thermotogae and it forms a Bacterial systematics monophyletic group with Coprothermobacter proteolyticus (a constituent of the phylum Firmicutes) and Coprothermobacter proteolyticus Thermotogae. Our findings indicate that C. proteolyticus does not belong to the phylum Firmicutes and that the Thermotogae phylum Dictyoglomi is not closely related to either the phylum Firmicutes or Synergistetes but to the phylum Thermotogae. © 2011 Elsevier Inc. -

The Ocean As a Global Reservoir of Antibiotic Resistance Genes
The Ocean as a Global Reservoir of Antibiotic Resistance Genes Stephen M. Hatosy,a Adam C. Martinya,b Department of Ecology and Evolutionary Biology, University of California, Irvine, California, USAa; Department of Earth System Science, University of California, Irvine, California, USAb Recent studies of natural environments have revealed vast genetic reservoirs of antibiotic resistance (AR) genes. Soil bacteria and Downloaded from human pathogens share AR genes, and AR genes have been discovered in a variety of habitats. However, there is little knowledge about the presence and diversity of AR genes in marine environments and which organisms host AR genes. To address this, we identified the diversity of genes conferring resistance to ampicillin, tetracycline, nitrofurantoin, and sulfadimethoxine in diverse marine environments using functional metagenomics (the cloning and screening of random DNA fragments). Marine environ- ments were host to a diversity of AR-conferring genes. Antibiotic-resistant clones were found at all sites, with 28% of the genes identified as known AR genes (encoding beta-lactamases, bicyclomycin resistance pumps, etc.). However, the majority of AR genes were not previously classified as such but had products similar to proteins such as transport pumps, oxidoreductases, and hydrolases. Furthermore, 44% of the genes conferring antibiotic resistance were found in abundant marine taxa (e.g., Pelagibac- http://aem.asm.org/ ter, Prochlorococcus, and Vibrio). Therefore, we uncovered a previously unknown diversity of genes that conferred an AR pheno- type among marine environments, which makes the ocean a global reservoir of both clinically relevant and potentially novel AR genes. he spread of antibiotic resistance (AR) is critically important sity of marine AR genes, and (iii) are these genes harbored by Tto human health. -

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Upper and Lower Case Letters to Be Used
Isolation, characterization and genome sequencing of a soil-borne Citrobacter freundii strain capable of detoxifying trichothecene mycotoxins by Rafiqul Islam A Thesis Presented to The University of Guelph In Partial Fulfilment of Requirements for the Degree of Doctor of Philosophy in Plant Agriculture Guelph, Ontario, Canada © Rafiqul Islam, April, 2012 ABSTRACT ISOLATION, CHARACTERIZATION AND GENOME SEQUENCING OF A SOIL- BORNE CITROBACTER FREUNDII STRAIN CAPABLE OF DETOXIFIYING TRICHOTHECENE MYCOTOXINS Rafiqul Islam Advisors: University of Guelph, 2012 Dr. K. Peter Pauls Dr. Ting Zhou Cereals are frequently contaminated with tricthothecene mycotoxins, like deoxynivalenol (DON, vomitoxin), which are toxic to humans, animals and plants. The goals of the research were to discover and characterize microbes capable of detoxifying DON under aerobic conditions and moderate temperatures. To identify microbes capable of detoxifying DON, five soil samples collected from Southern Ontario crop fields were tested for the ability to convert DON to a de-epoxidized derivative. One soil sample showed DON de-epoxidation activity under aerobic conditions at 22-24°C. To isolate the microbes responsible for DON detoxification (de-epoxidation) activity, the mixed culture was grown with antibiotics at 50ºC for 1.5 h and high concentrations of DON. The treatments resulted in the isolation of a pure DON de-epoxidating bacterial strain, ADS47, and phenotypic and molecular analyses identified the bacterium as Citrobacter freundii. The bacterium was also able to de-epoxidize and/or de-acetylate 10 other food-contaminating trichothecene mycotoxins. A fosmid genomic DNA library of strain ADS47 was prepared in E. coli and screened for DON detoxification activity. However, no library clone was found with DON detoxification activity. -

Antibiotic Resistance Determinant-Focused Acinetobacter Baumannii Vaccine Designed Using Reverse Vaccinology
International Journal of Molecular Sciences Article Antibiotic Resistance Determinant-Focused Acinetobacter baumannii Vaccine Designed Using Reverse Vaccinology Zhaohui Ni 1,2,†, Yan Chen 3,†, Edison Ong 2,4 and Yongqun He 2,5,* 1 Department of Pathogenobiology, College of Basic Medical Science, Jilin University, Changchun 130021, China; [email protected] 2 Unit for Laboratory Animal Medicine, Department of Microbiology and Immunology, University of Michigan, Ann Arbor, MI 48109, USA; [email protected] 3 Department of Neurosurgery, The Second Hospital of Jilin University, Changchun 130041, China; [email protected] 4 Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA 5 Center of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA * Correspondence: [email protected]; Tel.: +1-734-615-8231 † These authors contributed equally to this work. Academic Editor: Christopher Woelk Received: 27 December 2016; Accepted: 10 February 2017; Published: 21 February 2017 Abstract: As one of the most influential and troublesome human pathogens, Acinetobacter baumannii (A. baumannii) has emerged with many multidrug-resistant strains. After collecting 33 complete A. baumannii genomes and 84 representative antibiotic resistance determinants, we used the Vaxign reverse vaccinology approach to predict classical type vaccine candidates against A. baumannii infections and new type vaccine candidates against antibiotic resistance. Our genome analysis identified 35 outer membrane or extracellular adhesins that are conserved among all 33 genomes, have no human protein homology, and have less than 2 transmembrane helices. These 35 antigens include 11 TonB dependent receptors, 8 porins, 7 efflux pump proteins, and 2 fimbrial proteins (FilF and CAM87009.1). -

Xiexin Tang Improves the Symptom of Type 2 Diabetic Rats by Modulation of the Gut Microbiota
www.nature.com/scientificreports OPEN Xiexin Tang improves the symptom of type 2 diabetic rats by modulation of the gut microbiota Received: 30 August 2017 Xiaoyan Wei, Jinhua Tao , Suwei Xiao, Shu Jiang, Erxin Shang, Zhenhua Zhu, Dawei Qian Accepted: 13 February 2018 & Jinao Duan Published: xx xx xxxx Type 2 diabetes mellitus (T2DM), a chronic metabolic disease which severely impairs peoples’ quality of life, currently attracted worldwide concerns. There are growing evidences that gut microbiota can exert a great impact on the development of T2DM. Xiexin Tang (XXT), a traditional Chinese medicine prescription, has been clinically used to treat diabetes for thousands of years. However, few researches are investigated on the modulation of gut microbiota community by XXT which will be very helpful to unravel how it works. In this study, bacterial communities were analyzed based on high-throughput 16S rRNA gene sequencing. Results indicated that XXT could notably shape the gut microbiota. T2DM rats treated with XXT exhibited obvious changes in the composition of the gut microbiota, especially for some short chain fatty acids producing and anti-infammatory bacteria such as Adlercreutzia, Alloprevotella, Barnesiella, [Eubacterium] Ventriosum group, Blautia, Lachnospiraceae UCG-001, Papillibacter and Prevotellaceae NK3B31 group. Additionally, XXT could also signifcantly ameliorate hyperglycemia, lipid metabolism dysfunction and infammation in T2DM rats. Moreover, the correlation analysis illustrated that the key microbiota had a close relationship with the T2DM related indexes. The results probably provided useful information for further investigation on its active mechanism and clinical application. T2DM, a chronic metabolic disease characterized by hyperglycemia as a result of insufcient insulin secretion, insulin action or both1, is estimated that its numbers in the adults will increase by 55% by 20352. -

Table S4. Phylogenetic Distribution of Bacterial and Archaea Genomes in Groups A, B, C, D, and X
Table S4. Phylogenetic distribution of bacterial and archaea genomes in groups A, B, C, D, and X. Group A a: Total number of genomes in the taxon b: Number of group A genomes in the taxon c: Percentage of group A genomes in the taxon a b c cellular organisms 5007 2974 59.4 |__ Bacteria 4769 2935 61.5 | |__ Proteobacteria 1854 1570 84.7 | | |__ Gammaproteobacteria 711 631 88.7 | | | |__ Enterobacterales 112 97 86.6 | | | | |__ Enterobacteriaceae 41 32 78.0 | | | | | |__ unclassified Enterobacteriaceae 13 7 53.8 | | | | |__ Erwiniaceae 30 28 93.3 | | | | | |__ Erwinia 10 10 100.0 | | | | | |__ Buchnera 8 8 100.0 | | | | | | |__ Buchnera aphidicola 8 8 100.0 | | | | | |__ Pantoea 8 8 100.0 | | | | |__ Yersiniaceae 14 14 100.0 | | | | | |__ Serratia 8 8 100.0 | | | | |__ Morganellaceae 13 10 76.9 | | | | |__ Pectobacteriaceae 8 8 100.0 | | | |__ Alteromonadales 94 94 100.0 | | | | |__ Alteromonadaceae 34 34 100.0 | | | | | |__ Marinobacter 12 12 100.0 | | | | |__ Shewanellaceae 17 17 100.0 | | | | | |__ Shewanella 17 17 100.0 | | | | |__ Pseudoalteromonadaceae 16 16 100.0 | | | | | |__ Pseudoalteromonas 15 15 100.0 | | | | |__ Idiomarinaceae 9 9 100.0 | | | | | |__ Idiomarina 9 9 100.0 | | | | |__ Colwelliaceae 6 6 100.0 | | | |__ Pseudomonadales 81 81 100.0 | | | | |__ Moraxellaceae 41 41 100.0 | | | | | |__ Acinetobacter 25 25 100.0 | | | | | |__ Psychrobacter 8 8 100.0 | | | | | |__ Moraxella 6 6 100.0 | | | | |__ Pseudomonadaceae 40 40 100.0 | | | | | |__ Pseudomonas 38 38 100.0 | | | |__ Oceanospirillales 73 72 98.6 | | | | |__ Oceanospirillaceae -

Gut Microbiome Composition Remains Stable in Individuals with Diabetes-Related Early to Late Stage Chronic Kidney Disease
biomedicines Article Gut Microbiome Composition Remains Stable in Individuals with Diabetes-Related Early to Late Stage Chronic Kidney Disease Ashani Lecamwasam 1,2,3,*, Tiffanie M. Nelson 4, Leni Rivera 3, Elif I. Ekinci 2,5, Richard Saffery 1,6 and Karen M. Dwyer 3 1 Epigenetics Research, Murdoch Children’s Research Institute, VIC 3052, Australia; [email protected] 2 Department of Endocrinology, Austin Health, VIC 3079, Australia; [email protected] 3 School of Medicine, Faculty of Health, Deakin University, VIC 3220, Australia; [email protected] (L.R.); [email protected] (K.M.D.) 4 Menzies Health Institute Queensland, Griffith University, QLD 4222, Australia; [email protected] 5 Department of Medicine, University of Melbourne, VIC 3010, Australia 6 Department of Paediatrics, University of Melbourne, VIC 3010, Australia * Correspondence: [email protected]; Tel.: +613-8341-6200; Fax: +613-9348-1391 Abstract: (1) Background: Individuals with diabetes and chronic kidney disease display gut dysbiosis when compared to healthy controls. However, it is unknown whether there is a change in dysbiosis across the stages of diabetic chronic kidney disease. We investigated a cross-sectional study of patients with early and late diabetes associated chronic kidney disease to identify possible microbial differences between these two groups and across each of the stages of diabetic chronic kidney disease. (2) Methods: This cross-sectional study recruited 95 adults. DNA extracted from collected stool samples were used for 16S rRNA sequencing to identify the bacterial community in the Citation: Lecamwasam, A.; Nelson, gut. (3) Results: The phylum Firmicutes was the most abundant and its mean relative abundance T.M.; Rivera, L.; Ekinci, E.I.; Saffery, was similar in the early and late chronic kidney disease group, 45.99 ± 0.58% and 49.39 ± 0.55%, R.; Dwyer, K.M. -

Genus Contributing to the Top 70% of Significant Dissimilarity of Bacteria Between Day 7 and 18 Age Groups As Determined by SIMPER
Table S1: Genus contributing to the top 70% of significant dissimilarity of bacteria between day 7 and 18 age groups as determined by SIMPER. Overall average dissimilarity between ages is 51%. Day 7 Day 18 Average Average Phyla Class Order Family Genus % Abundance Abundance Actinobacteria Actinobacteria Actinomycetales Actinomycetaceae Actinomyces 0.57 0.24 0.74 Coriobacteriia Coriobacteriales Coriobacteriaceae Collinsella 0.51 0.61 0.57 Bacteroidetes Bacteroidia Bacteroidales Bacteroidaceae Bacteroides 2.1 1.63 1.03 Marinifilaceae Butyricimonas 0.82 0.81 0.9 Sanguibacteroides 0.08 0.42 0.59 CAG-873 0.01 1.1 1.68 Marinifilaceae 0.38 0.78 0.91 Marinifilaceae 0.78 1.04 0.97 p-2534-18B5 gut group 0.06 0.7 1.04 Prevotellaceae Alloprevotella 0.46 0.83 0.89 Prevotella 2 0.95 1.11 1.3 Prevotella 7 0.22 0.42 0.56 Prevotellaceae NK3B31 group 0.62 0.68 0.77 Prevotellaceae UCG-003 0.26 0.64 0.89 Prevotellaceae UCG-004 0.11 0.48 0.68 Prevotellaceae 0.64 0.52 0.89 Prevotellaceae 0.27 0.42 0.55 Rikenellaceae Alistipes 0.39 0.62 0.77 dgA-11 gut group 0.04 0.38 0.56 RC9 gut group 0.79 1.17 0.95 Epsilonbacteraeota Campylobacteria Campylobacterales Campylobacteraceae Campylobacter 0.58 0.83 0.97 Helicobacteraceae Helicobacter 0.12 0.41 0.6 Firmicutes Bacilli Lactobacillales Enterococcaceae Enterococcus 0.36 0.31 0.64 Lactobacillaceae Lactobacillus 1.4 1.24 1 Streptococcaceae Streptococcus 0.82 0.58 0.53 Firmicutes Clostridia Clostridiales Christensenellaceae Christensenellaceae R-7 group 0.38 1.36 1.56 Clostridiaceae Clostridium sensu stricto 1 1.16 0.58 -

Common Commensals
Common Commensals Actinobacterium meyeri Aerococcus urinaeequi Arthrobacter nicotinovorans Actinomyces Aerococcus urinaehominis Arthrobacter nitroguajacolicus Actinomyces bernardiae Aerococcus viridans Arthrobacter oryzae Actinomyces bovis Alpha‐hemolytic Streptococcus, not S pneumoniae Arthrobacter oxydans Actinomyces cardiffensis Arachnia propionica Arthrobacter pascens Actinomyces dentalis Arcanobacterium Arthrobacter polychromogenes Actinomyces dentocariosus Arcanobacterium bernardiae Arthrobacter protophormiae Actinomyces DO8 Arcanobacterium haemolyticum Arthrobacter psychrolactophilus Actinomyces europaeus Arcanobacterium pluranimalium Arthrobacter psychrophenolicus Actinomyces funkei Arcanobacterium pyogenes Arthrobacter ramosus Actinomyces georgiae Arthrobacter Arthrobacter rhombi Actinomyces gerencseriae Arthrobacter agilis Arthrobacter roseus Actinomyces gerenseriae Arthrobacter albus Arthrobacter russicus Actinomyces graevenitzii Arthrobacter arilaitensis Arthrobacter scleromae Actinomyces hongkongensis Arthrobacter astrocyaneus Arthrobacter sulfonivorans Actinomyces israelii Arthrobacter atrocyaneus Arthrobacter sulfureus Actinomyces israelii serotype II Arthrobacter aurescens Arthrobacter uratoxydans Actinomyces meyeri Arthrobacter bergerei Arthrobacter ureafaciens Actinomyces naeslundii Arthrobacter chlorophenolicus Arthrobacter variabilis Actinomyces nasicola Arthrobacter citreus Arthrobacter viscosus Actinomyces neuii Arthrobacter creatinolyticus Arthrobacter woluwensis Actinomyces odontolyticus Arthrobacter crystallopoietes -
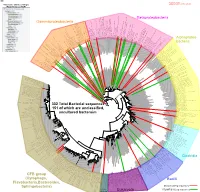
Bacteria Clostridia Bacilli Eukaryota CFB Group
AM935842.1.1361 uncultured Burkholderiales bacterium Class Betaproteobacteria AY283260.1.1552 Alcaligenes sp. PCNB−2 Class Betaproteobacteria AM934953.1.1374 uncultured Burkholderiales bacterium Class Betaproteobacteria AJ581593.1.1460 uncultured betaAM936569.1.1351 proteobacterium uncultured Class Betaproteobacteria Derxia sp. Class Betaproteobacteria AJ581621.1.1418 uncultured beta proteobacterium Class Betaproteobacteria DQ248272.1.1498 uncultured soil bacterium soil uncultured DQ248272.1.1498 DQ248235.1.1498 uncultured soil bacterium RS49 DQ248270.1.1496 uncultured soil bacterium DQ256489.1.1211 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax DQ256489.1.1211 AF523053.1.1486 uncultured Comamonadaceae bacterium Class Betaproteobacteria AY706442.1.1396 uncultured bacterium uncultured AY706442.1.1396 AJ536763.1.1422 uncultured bacterium CS000359.1.1530 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax CS000359.1.1530 AY168733.1.1411 uncultured bacterium AJ009470.1.1526 uncultured bacterium SJA−62 Class Betaproteobacteria Class SJA−62 bacterium uncultured AJ009470.1.1526 AY212561.1.1433 uncultured bacterium D16212.1.1457 Rhodoferax fermentans Class Betaproteobacteria Class fermentans Rhodoferax D16212.1.1457 AY957894.1.1546 uncultured bacterium AJ581620.1.1452 uncultured beta proteobacterium Class Betaproteobacteria RS76 AY625146.1.1498 uncultured bacterium RS65 DQ316832.1.1269 uncultured beta proteobacterium Class Betaproteobacteria DQ404909.1.1513 uncultured bacterium uncultured DQ404909.1.1513 AB021341.1.1466 bacterium rM6 AJ487020.1.1500 uncultured bacterium uncultured AJ487020.1.1500 RS7 RS86RC AF364862.1.1425 bacterium BA128 Class Betaproteobacteria AY957931.1.1529 uncultured bacterium uncultured AY957931.1.1529 CP000884.723807.725332 Delftia acidovorans SPH−1 Class Betaproteobacteria AY957923.1.1520 uncultured bacterium uncultured AY957923.1.1520 RS18 AY957918.1.1527 uncultured bacterium uncultured AY957918.1.1527 AY945883.1.1500 uncultured bacterium AF526940.1.1489 uncultured Ralstonia sp.