Diversity and Evolution of the Mhc-DRB1 Gene in the Two Endemic Iberian Subspecies of Pyrenean Chamois, Rupicapra Pyrenaica
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Contributions to the 12Th Conference of the European Wildlife Disease Association (EWDA) August 27Th – 31St, 2016, Berlin
12th Conference of the European Wildlife Disease Association (EWDA), Berlin 2016 Contributions to the 12th Conference of the European Wildlife Disease Association (EWDA) August 27th – 31st, 2016, Berlin, Germany Edited by Anke Schumann, Gudrun Wibbelt, Alex D. Greenwood, Heribert Hofer Organised by Leibniz Institute for Zoo and Wildlife Research (IZW) Alfred-Kowalke-Straße 17 10315 Berlin Germany www.izw-berlin.de and the European Wildlife Disease Association (EWDA) https://sites.google.com/site/ewdawebsite/ & ISBN 978-3-9815637-3-3 12th Conference of the European Wildlife Disease Association (EWDA), Berlin 2016 Published by Leibniz Institute for Zoo and Wildlife Research (IZW) Alfred-Kowalke-Str. 17, 10315 Berlin (Friedrichsfelde) PO Box 700430, 10324 Berlin, Germany Supported by Deutsche Forschungsgemeinschaft (DFG) [German Research Foundation] Kennedyallee 40, 53175 Bonn, Germany Printed on Forest Stewardship Council certified paper All rights reserved, particularly those for translation into other languages. It is not permitted to reproduce any part of this book by photocopy, microfilm, internet or any other means without written permission of the IZW. The use of product names, trade names or other registered entities in this book does not justify the assumption that these can be freely used by everyone. They may represent registered trademarks or other legal entities even if they are not marked as such. Processing of abstracts: Anke Schumann, Gudrun Wibbelt Setting and layout: Anke Schumann, Gudrun Wibbelt Cover: Diego Romero, Steven Seet, Gudrun Wibbelt Word cloud: ©Tagul.com Printing: Spree Druck Berlin GmbH www.spreedruck.de Order: Leibniz Institute for Zoo and Wildlife Research (IZW) Forschungsverbund Berlin e.V. PO Box 700430, 10324 Berlin, Germany [email protected] www.izw-berlin.de 12th Conference of the European Wildlife Disease Association (EWDA), Berlin 2016 CONTENTS Foreword ................................................................................................................. -
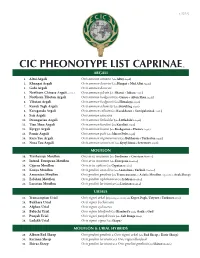
Cic Pheonotype List Caprinae©
v. 5.25.12 CIC PHEONOTYPE LIST CAPRINAE © ARGALI 1. Altai Argali Ovis ammon ammon (aka Altay Argali) 2. Khangai Argali Ovis ammon darwini (aka Hangai & Mid Altai Argali) 3. Gobi Argali Ovis ammon darwini 4. Northern Chinese Argali - extinct Ovis ammon jubata (aka Shansi & Jubata Argali) 5. Northern Tibetan Argali Ovis ammon hodgsonii (aka Gansu & Altun Shan Argali) 6. Tibetan Argali Ovis ammon hodgsonii (aka Himalaya Argali) 7. Kuruk Tagh Argali Ovis ammon adametzi (aka Kuruktag Argali) 8. Karaganda Argali Ovis ammon collium (aka Kazakhstan & Semipalatinsk Argali) 9. Sair Argali Ovis ammon sairensis 10. Dzungarian Argali Ovis ammon littledalei (aka Littledale’s Argali) 11. Tian Shan Argali Ovis ammon karelini (aka Karelini Argali) 12. Kyrgyz Argali Ovis ammon humei (aka Kashgarian & Hume’s Argali) 13. Pamir Argali Ovis ammon polii (aka Marco Polo Argali) 14. Kara Tau Argali Ovis ammon nigrimontana (aka Bukharan & Turkestan Argali) 15. Nura Tau Argali Ovis ammon severtzovi (aka Kyzyl Kum & Severtzov Argali) MOUFLON 16. Tyrrhenian Mouflon Ovis aries musimon (aka Sardinian & Corsican Mouflon) 17. Introd. European Mouflon Ovis aries musimon (aka European Mouflon) 18. Cyprus Mouflon Ovis aries ophion (aka Cyprian Mouflon) 19. Konya Mouflon Ovis gmelini anatolica (aka Anatolian & Turkish Mouflon) 20. Armenian Mouflon Ovis gmelini gmelinii (aka Transcaucasus or Asiatic Mouflon, regionally as Arak Sheep) 21. Esfahan Mouflon Ovis gmelini isphahanica (aka Isfahan Mouflon) 22. Larestan Mouflon Ovis gmelini laristanica (aka Laristan Mouflon) URIALS 23. Transcaspian Urial Ovis vignei arkal (Depending on locality aka Kopet Dagh, Ustyurt & Turkmen Urial) 24. Bukhara Urial Ovis vignei bocharensis 25. Afghan Urial Ovis vignei cycloceros 26. -

Low Sequence Diversity of the Prion Protein Gene (PRNP) in Wild Deer and Goat Species from Spain
Pitarch et al. Vet Res (2018) 49:33 https://doi.org/10.1186/s13567-018-0528-8 RESEARCH ARTICLE Open Access Low sequence diversity of the prion protein gene (PRNP) in wild deer and goat species from Spain José Luis Pitarch1, Helen Caroline Raksa1, María Cruz Arnal2, Miguel Revilla2, David Martínez2, Daniel Fernández de Luco2, Juan José Badiola1, Wilfred Goldmann3 and Cristina Acín1* Abstract The frst European cases of chronic wasting disease (CWD) in free-ranging reindeer and wild elk were confrmed in Norway in 2016 highlighting the urgent need to understand transmissible spongiform encephalopathies (TSEs) in the context of European deer species and the many individual populations throughout the European continent. The genetics of the prion protein gene (PRNP) are crucial in determining the relative susceptibility to TSEs. To establish PRNP gene sequence diversity for free-ranging ruminants in the Northeast of Spain, the open reading frame was sequenced in over 350 samples from fve species: Iberian red deer (Cervus elaphus hispanicus), roe deer (Capreolus capreolus), fallow deer (Dama dama), Iberian wild goat (Capra pyrenaica hispanica) and Pyrenean chamois (Rupicapra p. pyrenaica). Three single nucleotide polymorphisms (SNPs) were found in red deer: a silent mutation at codon 136, and amino acid changes T98A and Q226E. Pyrenean chamois revealed a silent SNP at codon 38 and an allele with a single octapeptide-repeat deletion. No polymorphisms were found in roe deer, fallow deer and Iberian wild goat. This appar- ently low variability of the PRNP coding region sequences of four major species in Spain resembles previous fndings for wild mammals, but implies that larger surveys will be necessary to fnd novel, low frequency PRNP gene alleles that may be utilized in CWD risk control. -

The Pyrenean Chamois Johan Espunyes Nozières
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Effects of global change on the diet of a mountain ungulate: the Pyrenean chamois Author Johan Espunyes Nozières Supervisors Emmanuel Serrano Ferron Mathieu Garel Oscar Cabezón Ponsoda Tutor Ignasi Marco Sánchez A dissertation for the degree of doctor philosophiae Departament de Medicina i Cirurgia Animals Facultat de Veterinària Universitat Autònoma de Barcelona 2019 1 This research was partially funded by the research partnership programme “Approche Intégrée de la Démographie des Populations d’Isard” (Nº2014/08/6171) between the Office de la Chasse et de la Faune Sauvage (ONCFS) and the Servei d’Ecopatologia de la Fauna Salvatge (SEFaS). Johan Espunyes Nozières acknowledges the Government of Andorra for a predoctoral grant, ATC015-AND-2015/2016, 2016/2017 and 2017/2018, a mobility grant AM059- AND-2018 and the award that covered the tuition fees of the third cycle studies AMTC0068-AND/2018. 2 Els doctors Emmanuel Serrano -

Spatial Patterns of Mitochondrial and Nuclear Gene Pools in Chamois (Rupicapra R
Heredity (2003) 91, 125–135 & 2003 Nature Publishing Group All rights reserved 0018-067X/03 $25.00 www.nature.com/hdy Spatial patterns of mitochondrial and nuclear gene pools in chamois (Rupicapra r. rupicapra) from the Eastern Alps H Schaschl, D Kaulfus, S Hammer1 and F Suchentrunk Research Institute of Wildlife Ecology, Veterinary Medicine University of Vienna, Savoyenstrasse 1, A-1160 Vienna, Austria We have assessed the variability of maternally (mtDNA) and variability as a result of immigration of chamois from different biparentally (allozymes) inherited genes of 443 chamois Pleistocene refugia surrounding the Alps after the withdrawal (Rupicapra r. rupicapra) from 19 regional samples in the of glaciers, rather than from topographic barriers to gene Eastern Alps, to estimate the degree and patterns of spatial flow, such as Alpine valleys, extended glaciers or woodlands. gene pool differentiation, and their possible causes. Based However, this striking geographical structuring of the on a total mtDNA-RFLP approach with 16 hexanucleotide- maternal genome was not paralleled by allelic variation at recognizing restriction endonucleases, we found marked 33 allozyme loci, which were used as nuclear DNA markers. substructuring of the maternal gene pool into four phylogeo- Wright’s hierarchical F-statistics revealed that only p0.45% of graphic groups. A hierarchical AMOVA revealed that 67.09% the explained allozymic diversity was because of partitioning of the variance was partitioned among these four mtDNA- among the four mtDNA-phylogroups. We conclude that this phylogroups, whereas only 8.04% were because of partition- discordance of spatial patterns of nuclear and mtDNA gene ing among regional samples within the populations, and pools results from a phylogeographic background and sex- 24.86% due to partitioning among individuals within regional specific dispersal, with higher levels of philopatry in females. -

6.5 X 11 Double Line.P65
Cambridge University Press 978-0-521-76059-1 - Ungulate Management in Europe: Problems and Practices Edited by Rory Putman, Marco Apollonio and Reidar Andersen Index More information Index adaptive management 134–5, 180–2, 377–9 chronic wasting disease 203 Alces alces – see moose climate change Ammotragus lervia – see Barbary sheep effects on ungulate populations 349–66 Axis axis – see axis deer effects on disease and disease transmission 203, axis deer 13, 35, 36 320, 335, 337–40, 360–1 conservation 4, 19, 26, 30, 32, 33, 39–41, 380 Barbary sheep 32, 35, 36 cultural attitudes to hunting 4, 5–9 Bison bonasus – see bison, European bison, European 15, 27–9, 32, 33, 38, 40, 269–70, Dama dama – see fallow deer 275, 301, 353 damage and its management 144–82 blue tongue virus 195, 199, 205, 208, 333, 338–9 damage to agricultural crops 144, 151, 169–71 boar, European wild 15, 81, 89, 91, 92, 93, 94–5, damage to forestry 35, 144, 149–50, 151, 96, 99, 117, 126, 129, 144, 170–1, 175, 200, 171–3 288, 292–4, 300, 301, 303, 322, 327, 334, 335, damage to conservation habitats 13, 35, 37, 43, 336, 355, 365 144, 173–5 bovine tuberculosis 194, 196, 200–1, 208, 322, damage, compensation for 68, 70–1, 169, 171 327, 334 damage, control of 176 brucellosis 197, 202, 208, 327 deer–vehicle collisions – see ungulate–vehicle collisions Capra aegagrus – see goat, European wild disease 192–209, 319–37 Capra ibex – see ibex, alpine disease surveillance 130, 203–4, 328–30 Capra pyrenaica – see ibex, Spanish ungulates as vectors for diseases of livestock or Capreolus -

Haplogroup Relationships Between Domestic and Wild Sheep Resolved Using a Mitogenome Panel
Heredity (2011) 106, 700–706 & 2011 Macmillan Publishers Limited All rights reserved 0018-067X/11 www.nature.com/hdy ORIGINAL ARTICLE Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel JRS Meadows1,3, S Hiendleder2 and JW Kijas1 1CSIRO Livestock Industries, St Lucia, Queensland, Australia and 2School of Agriculture, Food and Wine & Research Centre for Reproductive Health, School of Animal and Veterinary Science, University of Adelaide, Roseworthy, South Australia, Australia Five haplogroups have been identified in domestic sheep 920 000±190 000 years ago based on protein coding through global surveys of mitochondrial (mt) sequence sequence. The utility of various mtDNA components to inform variation, however these group classifications are often the true relationship between sheep was also examined with based on small fragments of the complete mtDNA sequence; Bayesian, maximum likelihood and partitioned Bremmer sup- partial control region or the cytochrome B gene. This study port analyses. The control region was found to be the mtDNA presents the complete mitogenome from representatives of component, which contributed the highest amount of support to each haplogroup identified in domestic sheep, plus a sample the tree generated using the complete data set. This study of their wild relatives. Comparison of the sequence success- provides the nucleus of a mtDNA mitogenome panel, which can fully resolved the relationships between each haplogroup be used to assess additional mitogenomes and serve as a and provided insight into the relationship with wild sheep. reference set to evaluate small fragments of the mtDNA. The five haplogroups were characterised as branching Heredity (2011) 106, 700–706; doi:10.1038/hdy.2010.122; independently, a radiation that shared a common ancestor published online 13 October 2010 Keywords: Ovis aries; domestication; mitochondria; genome; diversity Introduction Pedrosa et al., 2005; Pereira et al., 2006; Tapio et al., 2006; Meadows et al., 2007). -

Temporal Variation in Foraging Activity and Grouping Patterns in a Mountain-Dwelling Herbivore Environmental and Endogenous
Behavioural Processes 167 (2019) 103909 Contents lists available at ScienceDirect Behavioural Processes journal homepage: www.elsevier.com/locate/behavproc Temporal variation in foraging activity and grouping patterns in a mountain-dwelling herbivore: Environmental and endogenous drivers T ⁎ Niccolò Fattorinia, , Claudia Brunettia, Carolina Baruzzia, Gianpasquale Chiatanteb, Sandro Lovaria,c, Francesco Ferrettia a Department of Life Sciences, University of Siena. Via P.A. Mattioli 4, 53100 Siena, Italy b Department of Earth and Environmental Sciences, University of Pavia, Via Ferrata 1, 27100 Pavia, Italy c Maremma Natural History Museum, Strada Corsini 5, 58100 Grosseto, Italy ARTICLE INFO ABSTRACT Keywords: In temperate ecosystems, seasonality influences animal behaviour. Food availability, weather, photoperiod and Seasonality endogenous factors relevant to the biological cycle of individuals have been shown as major drivers of temporal Foraging Activity changes in activity rhythms and group size/structure of herbivorous species. We evaluated how diurnal female Group size foraging activity and grouping patterns of a mountain herbivore, the Apennine chamois Rupicapra pyrenaica Rupicapra ornata, varied during a decreasing gradient of pasture availability along the summer-autumn progression Chamois (July–October), a crucial period for the life cycle of mountain ungulates. Mountain herbivores Females increased diurnal foraging activity, possibly because of constrains elicited by variation in environ- mental factors. Size of mixed groups did not vary, in contrast with the hypothesis that groups should be smaller when pasture availability is lower. Proportion of females in groups increased, possibly suggesting that they concentrated on patchily distributed nutritious forbs. Occurrence of yearlings in groups decreased, which may have depended on dispersal of chamois in this age class. -

Infection Studies with Chamois Border Disease Virus in Pyrenean Chamois, Sheep and Pig
INFECTION STUDIES WITH CHAMOIS BORDER DISEASE VIRUS IN PYRENEAN CHAMOIS, SHEEP AND PIG ÒSCAR CABEZÓN PONSODA Directores: Ignasi Marco Sánchez Joaquim Segalés i Coma Departament de Medicina i Cirurgia Animals Facultat de Veterinària Universitat Autònoma de Barcelona 2011 1 2 Los Doctores Ignasi MARCO SÁNCHEZ y Joaquim SEGALÉS I COMA, Profesores Titulares de Universidad de las Áreas de conocimiento de Medicina y Cirugía Animal y Sanidad y Anatomía Animal, respectivamente, de la Facultad de Veterinaria de la Universitat Autònoma de Barcelona, HACEN CONSTAR, Que la memoria titulada “INFECTION STUDIES WITH CHAMOIS BORDER DISEASE VIRUS IN PYRENEAN CHAMOIS, SHEEP AND PIG”, presentada por Òscar Cabezón Ponsoda para la obtención del grado de Doctor en Veterinaria por la Universitat Autònoma de Barcelona, ha sido realizada bajo nuestra dirección y, considerándola satisfactoriamente finalizada, autorizamos su presentación para que sea juzgada por la comisión correspondiente. Y para que conste a los efectos oportunos, firmamos el presente informe en Bellaterra, a 1 de Junio de 2011. Firmado: Ignasi Marco Sánchez Firmado: Joaquim Segalés i Coma 3 4 AGRADECIMIENTOS No es habitual empezar los agradecimientos con alguien que no sea tu director de tesis, pero debo hacerlo. Y es que tengo que agradecer a Santiago Lavín primero, los 5 años que llevo en el SEFaS, y segundo, por la posibilidad de haber realizado esta tesis doctoral. Gracias, Santiago, por tener el convencimiento de que iba a ser así. Sin duda, y también antes de mis dos directores de Tesis, tengo que mencionar a Rosa Rosell. A Rosa le tengo que agradecer todo. Ella ha sido y es el mayor aliado que he tenido en todos los trabajos realizados. -

Cognitive Inferences in Fossil Apes (Primates, Hominoidea): Does Encephalization Reflect Intelligence?
JASs Invited Reviews Journal of Anthropological Sciences Vol. 88 (2010), pp. 11-48 Cognitive inferences in fossil apes (Primates, Hominoidea): does encephalization reflect intelligence? David M. Alba Institut Català de Paleontologia, Universitat Autònoma de Barcelona. Edifici ICP, Campus de la UAB s/n, 08193 Cerdanyola del Vallès, Barcelona (Spain); Dipartimento di Scienze della Terra, Università degli Studi di Firenze. Via G. La Pira 4, 50121 Firenze (Italy); e-mail: [email protected] Summary – Paleobiological inferences on general cognitive abilities (intelligence) in fossil hominoids strongly rely on relative brain size or encephalization, computed by means of allometric residuals, quotients or constants. This has been criticized on the basis that it presumably fails to reflect the higher intelligence of great apes, and absolute brain size has been favored instead. Many problems of encephalization metrics stem from the decrease of allometric slopes towards lower taxonomic level, thus making it difficult to determine at what level encephalization metrics have biological meaning. Here, the hypothesis that encephalization can be used as a good neuroanatomical proxy for intelligence is tested at two different taxonomic levels. A significant correlation is found between intelligence and encephalization only at a lower taxonomic level, i.e. on the basis of a low allometric slope, irrespective of whether species data or independent contrasts are employed. This indicates that higher-level slopes, resulting from encephalization grade shifts between subgroups (including hylobatids vs. great apes), do not reflect functional equivalence, whereas lower-level metrics can be employed as a paleobiological proxy for intelligence. Thus, in accordance to intelligence rankings, lower-level metrics indicate that great apes are more encephalized than both monkeys and hylobatids. -

Nesiotites Sample
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Repositorio Universidad de Zaragoza Molecular phylogenetics supports the origin of an endemic Balearic shrew lineage (Nesiotites) coincident with the Messinian Salinity Crisis Pere Bovera,b,c*, Kieren J. Mitchella, Bastien Llamasa, Juan Rofesd, Vicki A. Thomsone, Gloria Cuenca-Bescósf, Josep A. Alcoverb,c, Alan Coopera, Joan Ponsb a Australian Centre for Ancient DNA (ACAD), School of Biological Sciences, University of Adelaide, Australia b Departament de Biodiversitat i Conservació, Institut Mediterrani d’Estudis Avançats (CSIC-UIB), Esporles, Illes Balears, Spain c Research Associate, Department of Mammalogy/Division of Vertebrate Zoology, American Museum of Natural History, NY d Archéozoologie, Archéobotanique: Sociétés, pratiques et environnements (UMR 7209), Sorbonne Universités, Muséum national d'Histoire naturelle, CNRS, CP56, 55 rue Buffon, 75005 Paris, France. e School of Biological Sciences, University of Adelaide, Australia. f Grupo Aragosaurus-IUCA, Universidad de Zaragoza, Spain. * Corresponding author at: Australian Centre for Ancient DNA (ACAD), School of Biological Sciences, University of Adelaide, Darling Building, North Terrace Campus, Adelaide, SA, 5005, Australia (P. Bover). E-mail addresses: [email protected] (P. Bover), [email protected] (K.J. Mitchell), [email protected] (B. Llamas), [email protected] (J. Rofes), [email protected] (V. Thomson), [email protected] (G. Cuenca-Bescós), [email protected] (J.A. Alcover), [email protected] (A. Cooper), [email protected] (J. Pons). Abstract The red-toothed shrews (Soricinae) are the most widespread subfamily of shrews, distributed from northern South America to North America and Eurasia. -

Brochure Highlight Those Impressive Russia
2019 44 years and counting The products and services listed Join us on Facebook, follow us on Instagram or visit our web site to become one Table of Contents in advertisements are offered and of our growing number of friends who receive regular email updates on conditions Alaska . 4 provided solely by the advertiser. and special big game hunt bargains. Australia . 38 www.facebook.com/NealAndBrownleeLLC Neal and Brownlee, L.L.C. offers Austria . 35 Instagram: @NealAndBrownleeLLC no guarantees, warranties or Azerbaijan . 31 recommendations for the services or Benin . 18 products offered. If you have questions Cameroon . 19 related to these services, please contact Canada . 6 the advertiser. Congo . 20 All prices, terms and conditions Continental U .S . 12 are, to the best of our knowledge at the Ethiopia . 20 time of printing, the most recent and Fishing Alaska . 42 accurate. Prices, terms and conditions Fishing British Columbia . 41 are subject to change without notice Fishing New Zealand . 42 due to circumstances beyond our Kyrgyzstan . 31 control. Jeff C. Neal Greg Brownlee Trey Sperring Mexico . 14 Adventure travel and big game 2018 was another fantastic year for our company thanks to the outfitters we epresentr and the Mongolia . 32 hunting contain inherent risks and clients who trusted us. We saw more clients traveling last season than in any season in the past, Mozambique . 21 dangers by their very nature that with outstanding results across the globe. African hunting remained strong, with our primary Namibia . 22 are beyond the control of Neal and areas producing outstanding success across several countries. Asian hunting has continued to be Nepal .