Haplogroup Relationships Between Domestic and Wild Sheep Resolved Using a Mitogenome Panel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Germanic Origins from the Perspective of the Y-Chromosome
Germanic Origins from the Perspective of the Y-Chromosome By Michael Robert St. Clair A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor in Philosophy in German in the Graduate Division of the University of California, Berkeley Committee in charge: Irmengard Rauch, Chair Thomas F. Shannon Montgomery Slatkin Spring 2012 Abstract Germanic Origins from the Perspective of the Y-Chromosome by Michael Robert St. Clair Doctor of Philosophy in German University of California, Berkeley Irmengard Rauch, Chair This dissertation holds that genetic data are a useful tool for evaluating contemporary models of Germanic origins. The Germanic languages are a branch of the Indo-European language family and include among their major contemporary representatives English, German, Dutch, Danish, Swedish, Norwegian and Icelandic. Historically, the search for Germanic origins has sought to determine where the Germanic languages evolved, and why the Germanic languages are similar to and different from other European languages. Both archaeological and linguist approaches have been employed in this research direction. The linguistic approach to Germanic origins is split among those who favor the Stammbaum theory and those favoring language contact theory. Stammbaum theory posits that Proto-Germanic separated from an ancestral Indo-European parent language. This theoretical approach accounts for similarities between Germanic and other Indo- European languages by posting a period of mutual development. Germanic innovations, on the other hand, occurred in isolation after separation from the parent language. Language contact theory posits that Proto-Germanic was the product of language convergence and this convergence explains features that Germanic shares with other Indo-European languages. -

Mitochondrial Haplogroup Background May Influence
Genetics Mitochondrial Haplogroup Background May Influence Southeast Asian G11778A Leber Hereditary Optic Neuropathy Supannee Kaewsutthi,1,2 Nopasak Phasukkijwatana,2,3 Yutthana Joyjinda,1 Wanicha Chuenkongkaew,3,4 Bussaraporn Kunhapan,1 Aung Win Tun,1 Bhoom Suktitipat,1 and Patcharee Lertrit1,4 PURPOSE. To investigate the role of mitochondrial DNA markedly incomplete penetrance. The three most common (mt DNA) background on the expression of Leber hereditary primary LHON mutations, G3460A in ND1, G11778A in ND4, optic neuropathy (LHON) in Southeast Asian carriers of the and T14484C in ND6, account for more than 90% of LHON G11778A mutation. cases worldwide2 with G11778A being the most common. In 3 4–6 METHODS. Complete mtDNA sequences were analyzed from 53 Thailand and other Asian countries, G11778A is responsi- unrelated Southeast Asian G11778A LHON pedigrees in Thai- ble for approximately 90% of LHON families. land and 105 normal Thai controls, and mtDNA haplogroups The sex bias and the marked incomplete penetrance of were determined. Clinical phenotypes were tested for associ- LHON indicate that there must be other factors that modify disease expression. Mitochondrial background,7–8 nuclear ation with mtDNA haplogroup, with adjustment for potential 9–11 12 confounders such as sex and age at onset. background, and environmental factors have been impli- cated in disease expression, although the precise mechanisms RESULTS. mtDNA subhaplogroup B was significantly associated of pathogenesis are largely undefined. with LHON. Follow-up analysis -

An Overview of the Independent Histories of the Human Y Chromosome and the Human Mitochondrial Chromosome
The Proceedings of the International Conference on Creationism Volume 8 Print Reference: Pages 133-151 Article 7 2018 An Overview of the Independent Histories of the Human Y Chromosome and the Human Mitochondrial chromosome Robert W. Carter Stephen Lee University of Idaho John C. Sanford Cornell University, Cornell University College of Agriculture and Life Sciences School of Integrative Plant Science,Follow this Plant and Biology additional Section works at: https://digitalcommons.cedarville.edu/icc_proceedings DigitalCommons@Cedarville provides a publication platform for fully open access journals, which means that all articles are available on the Internet to all users immediately upon publication. However, the opinions and sentiments expressed by the authors of articles published in our journals do not necessarily indicate the endorsement or reflect the views of DigitalCommons@Cedarville, the Centennial Library, or Cedarville University and its employees. The authors are solely responsible for the content of their work. Please address questions to [email protected]. Browse the contents of this volume of The Proceedings of the International Conference on Creationism. Recommended Citation Carter, R.W., S.S. Lee, and J.C. Sanford. An overview of the independent histories of the human Y- chromosome and the human mitochondrial chromosome. 2018. In Proceedings of the Eighth International Conference on Creationism, ed. J.H. Whitmore, pp. 133–151. Pittsburgh, Pennsylvania: Creation Science Fellowship. Carter, R.W., S.S. Lee, and J.C. Sanford. An overview of the independent histories of the human Y-chromosome and the human mitochondrial chromosome. 2018. In Proceedings of the Eighth International Conference on Creationism, ed. J.H. -

REPORT on TRANSBOUNDARY CONSERVATION HOTSPOTS for the CENTRAL ASIAN MAMMALS INITIATIVE (Prepared by the Secretariat)
CONVENTION ON UNEP/CMS/COP13/Inf.27 MIGRATORY 8 January 2020 SPECIES Original: English 13th MEETING OF THE CONFERENCE OF THE PARTIES Gandhinagar, India, 17 - 22 February 2020 Agenda Item 26.3 REPORT ON TRANSBOUNDARY CONSERVATION HOTSPOTS FOR THE CENTRAL ASIAN MAMMALS INITIATIVE (Prepared by the Secretariat) Summary: This report was developed with funding from the Government of Switzerland within the frame of the Central Asian Mammals Initiative (CAMI) (Doc. 26.3.5) to identify transboundary conservation hotspots and develop recommendations for their conservation. The report builds on existing projects, in particular, the CAMI Linear Infrastructure and Migration Atlas (see Inf.Doc.19) and focusses on the same species and geographical area. The study was discussed during the CAMI Range State Meeting held from 25-28 September 2019 in Ulaanbaatar, Mongolia where participants reviewed the pre-identified areas. Their comments are incorporated in this report. Participants also provided new information about important transboundary sites from Bhutan, India, Nepal and Pakistan and recommended to send the report for final review to Range States and experts. It was also recommended that the final report covers all CAMI species as adopted at COP13. This report is therefore a final draft with the last step to expand the geographical and species scope and finalize the report to be undertaken after COP13. Mapping Transboundary Conservation Hotspots for the Central Asian Mammals Initiative Photo credit: Viktor Lukarevsky Report – Draft 5 incorporating comments made during the CAMI Range States Meeting on 25-28 September 2019 in Ulaanbaatar, Mongolia. The report does not yet consider the Urial, Persian leopard and Gobi bear as CAMI species pending decision at the CMS COP13, as well as the proposed expansion of the geographic and species scope to include the entire CAMI region in this study. -
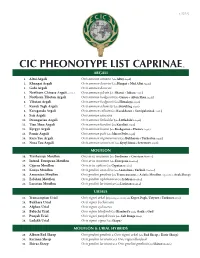
Cic Pheonotype List Caprinae©
v. 5.25.12 CIC PHEONOTYPE LIST CAPRINAE © ARGALI 1. Altai Argali Ovis ammon ammon (aka Altay Argali) 2. Khangai Argali Ovis ammon darwini (aka Hangai & Mid Altai Argali) 3. Gobi Argali Ovis ammon darwini 4. Northern Chinese Argali - extinct Ovis ammon jubata (aka Shansi & Jubata Argali) 5. Northern Tibetan Argali Ovis ammon hodgsonii (aka Gansu & Altun Shan Argali) 6. Tibetan Argali Ovis ammon hodgsonii (aka Himalaya Argali) 7. Kuruk Tagh Argali Ovis ammon adametzi (aka Kuruktag Argali) 8. Karaganda Argali Ovis ammon collium (aka Kazakhstan & Semipalatinsk Argali) 9. Sair Argali Ovis ammon sairensis 10. Dzungarian Argali Ovis ammon littledalei (aka Littledale’s Argali) 11. Tian Shan Argali Ovis ammon karelini (aka Karelini Argali) 12. Kyrgyz Argali Ovis ammon humei (aka Kashgarian & Hume’s Argali) 13. Pamir Argali Ovis ammon polii (aka Marco Polo Argali) 14. Kara Tau Argali Ovis ammon nigrimontana (aka Bukharan & Turkestan Argali) 15. Nura Tau Argali Ovis ammon severtzovi (aka Kyzyl Kum & Severtzov Argali) MOUFLON 16. Tyrrhenian Mouflon Ovis aries musimon (aka Sardinian & Corsican Mouflon) 17. Introd. European Mouflon Ovis aries musimon (aka European Mouflon) 18. Cyprus Mouflon Ovis aries ophion (aka Cyprian Mouflon) 19. Konya Mouflon Ovis gmelini anatolica (aka Anatolian & Turkish Mouflon) 20. Armenian Mouflon Ovis gmelini gmelinii (aka Transcaucasus or Asiatic Mouflon, regionally as Arak Sheep) 21. Esfahan Mouflon Ovis gmelini isphahanica (aka Isfahan Mouflon) 22. Larestan Mouflon Ovis gmelini laristanica (aka Laristan Mouflon) URIALS 23. Transcaspian Urial Ovis vignei arkal (Depending on locality aka Kopet Dagh, Ustyurt & Turkmen Urial) 24. Bukhara Urial Ovis vignei bocharensis 25. Afghan Urial Ovis vignei cycloceros 26. -

Different Matrilineal Contributions to Genetic Structure of Ethnic Groups in the Silk Road Region in China
Different Matrilineal Contributions to Genetic Structure of Ethnic Groups in the Silk Road Region in China Yong-Gang Yao,*1 Qing-Peng Kong,* à1 Cheng-Ye Wang,*à Chun-Ling Zhu,* and Ya-Ping Zhang* *Laboratory of Cellular and Molecular Evolution, and Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China; Laboratory for Conservation and Utilization of Bio-resource, Yunnan University, Kunming, China; and àGraduate School of the Chinese Academy of Sciences, Beijing, China Previous studies have shown that there were extensive genetic admixtures in the Silk Road region. In the present study, we analyzed 252 mtDNAs of five ethnic groups (Uygur, Uzbek, Kazak, Mongolian, and Hui) from Xinjiang Province, China (through which the Silk Road once ran) together with some reported data from the adjacent regions in Central Asia. In a simple way, we classified the mtDNAs into different haplogroups (monophyletic clades in the rooted mtDNA tree) according to the available phylogenetic information and compared their frequencies to show the differences among Downloaded from https://academic.oup.com/mbe/article/21/12/2265/1071048 by guest on 27 September 2021 the matrilineal genetic structures of these populations with different demographic histories. With the exception of eight unassigned M*, N*, and R* mtDNAs, all the mtDNA types identified here belonged to defined subhaplogroups of haplogroups M and N (including R) and consisted of subsets of both the eastern and western Eurasian pools, thus providing direct evidence supporting the suggestion that Central Asia is the location of genetic admixture of the East and the West. -

Early Survival of Punjab Urial
394 Early survival of Punjab urial Ghulam Ali Awan, Marco Festa-Bianchet, and Jean-Michel Gaillard Abstract: There is almost no information on age-specific survival of Asiatic ungulates based on mark–recapture studies. Survival of marked Punjab urial (Ovis vignei punjabiensis Lydekker, 1913) aged 0–2 years was studied in the Salt Range, Pakistan, in 2001–2005. Male lambs were heavier than females at birth. The relationship between litter size and birth mass varied among years, with a tendency for twins to be lighter than singletons. Birth mass had a positive but nonsignificant relation with survival to 1 year. Neither sex nor litter size affected survival to 1 year, which averaged 55% (95% CI = 41%–68%). There was no sex effect on survival of yearlings, which averaged 88% (95% CI = 4%–100%). Although sur- vival of lambs and yearlings was similar to that reported for other ungulates, apparent survival of 2- and 3-year-olds was very low at only 47%, possibly because of emigration. Early survival in this protected area is adequate to allow population growth, but more data are required on adult survival. Re´sume´ : En l’absence de suivis par capture–marquage–recapture, il n’y a pratiquement pas d’information disponible sur la variation des taux de survie en fonction de l’aˆge pour les espe`ces d’ongule´s asiatiques. Nous avons e´tudie´ la survie d’urials du Punjab (Ovis vignei punjabiensis Lydekker, 1913) individuellement marque´s entre 0 et 2 ans dans la re´gion de la chaıˆne de Salt, au Pakistan, entre 2001 et 2005. -

Status and Red List of Pakistan's Mammals
SSttaattuuss aanndd RReedd LLiisstt ooff PPaakkiissttaann’’ss MMaammmmaallss based on the Pakistan Mammal Conservation Assessment & Management Plan Workshop 18-22 August 2003 Authors, Participants of the C.A.M.P. Workshop Edited and Compiled by, Kashif M. Sheikh PhD and Sanjay Molur 1 Published by: IUCN- Pakistan Copyright: © IUCN Pakistan’s Biodiversity Programme This publication can be reproduced for educational and non-commercial purposes without prior permission from the copyright holder, provided the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited without prior permission (in writing) of the copyright holder. Citation: Sheikh, K. M. & Molur, S. 2004. (Eds.) Status and Red List of Pakistan’s Mammals. Based on the Conservation Assessment and Management Plan. 312pp. IUCN Pakistan Photo Credits: Z.B. Mirza, Kashif M. Sheikh, Arnab Roy, IUCN-MACP, WWF-Pakistan and www.wildlife.com Illustrations: Arnab Roy Official Correspondence Address: Biodiversity Programme IUCN- The World Conservation Union Pakistan 38, Street 86, G-6⁄3, Islamabad Pakistan Tel: 0092-51-2270686 Fax: 0092-51-2270688 Email: [email protected] URL: www.biodiversity.iucnp.org or http://202.38.53.58/biodiversity/redlist/mammals/index.htm 2 Status and Red List of Pakistan Mammals CONTENTS Contributors 05 Host, Organizers, Collaborators and Sponsors 06 List of Pakistan Mammals CAMP Participants 07 List of Contributors (with inputs on Biological Information Sheets only) 09 Participating Institutions -

Cervid Mixed-Species Table That Was Included in the 2014 Cervid RC
Appendix III. Cervid Mixed Species Attempts (Successful) Species Birds Ungulates Small Mammals Alces alces Trumpeter Swans Moose Axis axis Saurus Crane, Stanley Crane, Turkey, Sandhill Crane Sambar, Nilgai, Mouflon, Indian Rhino, Przewalski Horse, Sable, Gemsbok, Addax, Fallow Deer, Waterbuck, Persian Spotted Deer Goitered Gazelle, Reeves Muntjac, Blackbuck, Whitetailed deer Axis calamianensis Pronghorn, Bighorned Sheep Calamian Deer Axis kuhili Kuhl’s or Bawean Deer Axis porcinus Saurus Crane Sika, Sambar, Pere David's Deer, Wisent, Waterbuffalo, Muntjac Hog Deer Capreolus capreolus Western Roe Deer Cervus albirostris Urial, Markhor, Fallow Deer, MacNeil's Deer, Barbary Deer, Bactrian Wapiti, Wisent, Banteng, Sambar, Pere White-lipped Deer David's Deer, Sika Cervus alfredi Philipine Spotted Deer Cervus duvauceli Saurus Crane Mouflon, Goitered Gazelle, Axis Deer, Indian Rhino, Indian Muntjac, Sika, Nilgai, Sambar Barasingha Cervus elaphus Turkey, Roadrunner Sand Gazelle, Fallow Deer, White-lipped Deer, Axis Deer, Sika, Scimitar-horned Oryx, Addra Gazelle, Ankole, Red Deer or Elk Dromedary Camel, Bison, Pronghorn, Giraffe, Grant's Zebra, Wildebeest, Addax, Blesbok, Bontebok Cervus eldii Urial, Markhor, Sambar, Sika, Wisent, Waterbuffalo Burmese Brow-antlered Deer Cervus nippon Saurus Crane, Pheasant Mouflon, Urial, Markhor, Hog Deer, Sambar, Barasingha, Nilgai, Wisent, Pere David's Deer Sika 52 Cervus unicolor Mouflon, Urial, Markhor, Barasingha, Nilgai, Rusa, Sika, Indian Rhino Sambar Dama dama Rhea Llama, Tapirs European Fallow Deer -

Genetics, Linguistics, and Prehistoric Migrations: an Analysis of California Indian Mitochondrial DNA Lineages
Journal of California and Great Basin Anttiropology | Vol, 26, No, 1 (2006) | pp 33-64 Genetics, Linguistics, and Prehistoric Migrations: An Analysis of California Indian Mitochondrial DNA Lineages JOHN R. JOHNSON Department of Anthropology, Santa Barbara Museum of Natural History 2559 Puesta del Sol, Santa Barbara, CA 93105 JOSEPH G. LORENZ Laboratory of Molecular Biology, Coriell Institute for Medical Research 403 Haddon Avenue, Camden, NJ 08103 The advent of mitochondrial DNA analysis makes possible the study of past migrations among California Indians through the study of genetic similarities and differences. Four scenarios of language change correlate with observable genetic patterns: (1) initial colonization followed by gradual changes due to isolation; (2) population replacement; (3) elite dominance; and (4) intermarriage between adjacent groups. A total of 126 mtDNA samples were provided by contemporary California Indian descendants whose maternal lineages were traced back to original eighteenth and nineteenth century sociolinguistic groups using mission records and other ethnohistoric sources. In particular, those groups belonging to three language families (Chumashan, Uto-Aztecan, and Yokutsan) encompassed enough samples to make meaningful comparisons. The four predominant mtDNA haplogroups found among American Indians (A, B, C, and D) were distributed differently among populations belonging to these language families in California. Examination of the distribution of particular haplotypes within each haplogroup further elucidated the separate population histories of these three language families. The expansions of Yokutsan and Uto-Aztecan groups into their respective homelands are evident in the structure of genetic relationships within haplogroup diagrams. The ancient presence of Chumashan peoples in the Santa Barbara Channel region can be inferred from the presence of a number of haplotypes arrayed along a chain-like branch derived from the founding haplotype within Haplogroup A. -

Diversity and Evolution of the Mhc-DRB1 Gene in the Two Endemic Iberian Subspecies of Pyrenean Chamois, Rupicapra Pyrenaica
Heredity (2007) 99, 406–413 & 2007 Nature Publishing Group All rights reserved 0018-067X/07 $30.00 www.nature.com/hdy ORIGINAL ARTICLE Diversity and evolution of the Mhc-DRB1 gene in the two endemic Iberian subspecies of Pyrenean chamois, Rupicapra pyrenaica J Alvarez-Busto1, K Garcı´a-Etxebarria1, J Herrero2,3, I Garin4 and BM Jugo1 1Genetika, Antropologia Fisikoa eta Animali Fisiologia Saila, Zientzia eta Teknologia Fakultatea, Euskal Herriko Unibertsitatea (UPV/EHU), Bilbao, Spain; 2Departamento de Ecologı´a, Universidad de Alcala´ de Henares, Alcala´ de Henares, Spain; 3EGA Wildlife Consultants, Zaragoza, Spain and 4Zoologı´a eta Animali Zelulen Biologia Saila, Zientzia eta Teknologia Fakultatea, Euskal Herriko Unibertsitatea (UPV/EHU), Bilbao, Spain Major histocompatibility complex class II locus DRB variation recent parasitic infections by sarcoptic mange. A phyloge- was investigated by single-strand conformation polymorph- netic analysis of both Pyrenean chamois and DRB alleles ism analysis and sequence analysis in the two subspecies of from 10 different caprinid species revealed that the chamois Pyrenean chamois (Rupicapra pyrenaica) endemic to the alleles form two monophyletic groups. In comparison with Iberian Peninsula. Low levels of genetic variation were other Caprinae DRB sequences, the Rupicapra alleles detected in both subspecies, with seven different alleles in R. displayed a species-specific clustering that reflects a large p. pyrenaica and only three in the R. p. parva. After applying temporal divergence of the chamois from other caprinids, as the rarefaction method to cope with the differences in sample well as a possible difference in the selective environment for size, the low allele number of parva was highlighted. -

Using Ancient DNA to Study the Origins and Dispersal of Ancestral Polynesian Chickens Across the Pacific
Using ancient DNA to study the origins and dispersal of ancestral Polynesian chickens across the Pacific Vicki A. Thomsona, Ophélie Lebrasseurb, Jeremy J. Austina,c,1, Terry L. Huntd,e, David A. Burneyf, Tim Denhamg, Nicolas J. Rawlencea,h, Jamie R. Woodi, Jaime Gongoraj, Linus Girdland Flinkb,k, Anna Linderholmb, Keith Dobneyl, Greger Larsonb, and Alan Coopera,1 aAustralian Centre for Ancient DNA, School of Earth and Environmental Sciences, University of Adelaide, Adelaide, SA 5005, Australia; bDurham Evolution and Ancient DNA, Department of Archaeology, and kSchool of Biological and Biomedical Sciences, Durham University, Durham DH1 3LE, United Kingdom; cSciences Department, Museum Victoria, Melbourne, VIC 3001, Australia; dClark Honors College and eDepartment of Anthropology, University of Oregon, Eugene, OR 97403; fNational Tropical Botanical Garden, Kalaheo, HI 96741; gSchool of Archaeology and Anthropology, ANU College of Arts and Social Sciences, The Australian National University, Canberra, ACT 0200, Australia; hAllan Wilson Centre for Molecular Ecology and Evolution, Department of Zoology, University of Otago, Dunedin 9016, New Zealand; iLandcare Research, Lincoln 7640, New Zealand; jFaculty of Veterinary Science, University of Sydney, Sydney, NSW 2006, Australia; and lDepartment of Archaeology, University of Aberdeen, Aberdeen AB24 3UF, Scotland Edited by David J. Meltzer, Southern Methodist University, Dallas, TX, and approved February 20, 2014 (received for review October 31, 2013) The human colonization of Remote Oceania remains one of the resolved, the precise details of this intensive migratory episode great feats of exploration in history, proceeding east from Asia remain unclear (6). across the vast expanse of the Pacific Ocean. Human commensal Human commensal and early domesticated species were wide- and domesticated species were widely transported as part of this ly, but not ubiquitously, dispersed as people colonized the Pacific.