Phylogeny and Phylogenetic Nomenclature of the Campanulidae Based on an Expanded Sample of Genes and Taxa
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Toward a Resolution of Campanulid Phylogeny, with Special Reference to the Placement of Dipsacales
TAXON 57 (1) • February 2008: 53–65 Winkworth & al. • Campanulid phylogeny MOLECULAR PHYLOGENETICS Toward a resolution of Campanulid phylogeny, with special reference to the placement of Dipsacales Richard C. Winkworth1,2, Johannes Lundberg3 & Michael J. Donoghue4 1 Departamento de Botânica, Instituto de Biociências, Universidade de São Paulo, Caixa Postal 11461–CEP 05422-970, São Paulo, SP, Brazil. [email protected] (author for correspondence) 2 Current address: School of Biology, Chemistry, and Environmental Sciences, University of the South Pacific, Private Bag, Laucala Campus, Suva, Fiji 3 Department of Phanerogamic Botany, The Swedish Museum of Natural History, Box 50007, 104 05 Stockholm, Sweden 4 Department of Ecology & Evolutionary Biology and Peabody Museum of Natural History, Yale University, P.O. Box 208106, New Haven, Connecticut 06520-8106, U.S.A. Broad-scale phylogenetic analyses of the angiosperms and of the Asteridae have failed to confidently resolve relationships among the major lineages of the campanulid Asteridae (i.e., the euasterid II of APG II, 2003). To address this problem we assembled presently available sequences for a core set of 50 taxa, representing the diver- sity of the four largest lineages (Apiales, Aquifoliales, Asterales, Dipsacales) as well as the smaller “unplaced” groups (e.g., Bruniaceae, Paracryphiaceae, Columelliaceae). We constructed four data matrices for phylogenetic analysis: a chloroplast coding matrix (atpB, matK, ndhF, rbcL), a chloroplast non-coding matrix (rps16 intron, trnT-F region, trnV-atpE IGS), a combined chloroplast dataset (all seven chloroplast regions), and a combined genome matrix (seven chloroplast regions plus 18S and 26S rDNA). Bayesian analyses of these datasets using mixed substitution models produced often well-resolved and supported trees. -

The New Zealand Rain Forest: a Comparison with Tropical Rain Forest! J
The New Zealand Rain Forest: A Comparison with Tropical Rain Forest! J. W. DAWSON2 and B. V. SNEDDON2 ABSTRACT: The structure of and growth forms and habits exhibited by the New Zealand rain forest are described and compared with those of lowland tropical rain forest. Theories relating to the frequent regeneration failure of the forest dominants are outlined. The floristic affinities of the forest type are discussed and it is suggested that two main elements can be recognized-lowland tropical and montane tropical. It is concluded that the New Zealand rain forest is comparable to lowland tropical rain forest in structure and in range of special growth forms and habits. It chiefly differs in its lower stature, fewer species, and smaller leaves. The floristic similarity between the present forest and forest floras of the Tertiary in New Zealand suggest that the former may be a floristically reduced derivative of the latter. PART 1 OF THIS PAPER describes the structure The approximate number of species of seed and growth forms of the New Zealand rain plants in these forests is 240. From north to forest as exemplified by a forest in the far north. south there is an overall decrease in number of In Part 2, theories relating to the regeneration species. At about 38°S a number of species, of the dominant trees in the New Zealand rain mostly trees and shrubs, drop out or become forest generally are reviewed briefly, and their restricted to coastal sites, but it is not until about relevance to the situation in the study forest is 42°S, in the South Island, that many of the con considered. -

The Species of Alseuosmia (Alseuosmiaceae)
New Zealand Journal of Botany, 1978, Vol. 16: 271-7. 271 The species of Alseuosmia (Alseuosmiaceae) RHYS O. GARDNER Department of Botany, University of Auckland, Private Bag, Auckland, New Zealand (Received 15 September 1977) ABSTRACT A new species Alseuosmia turneri R. 0. Gardner (Alseuosmiaceae Airy Shaw) from the Volcanic Plateau, North Island, New Zealand is described and illustrated. A. linariifolia A. Cunn. is reduced to a variety of A. banksii A. Cunn. and A. quercifolia A. Cunn. is given hybrid status (=^4. banksii A. Cunn. x A. macrophylla A. Cunn.). A key to the four Alseuosmia species, synonymy, and a generalised distribution map are given. A. pusilla Col. is illustrated for the first time. INTRODUCTION judged to be frequent between only one pair of Allan Cunningham (1839) described eight species and the "excessive variability" lies mostly species of a new flowering plant genus Alseuosmia there. from material collected in the Bay of Islands region by Banks and Solander in 1769, by himself in 1826 and 1838, and by his brother Richard in 1833-4. The A NEW SPECIES OF ALSEUOSMIA species, all shrubs of the lowland forest, were sup- A. CUNN. posed to differ from one another chiefly in the shape and toothing of their leaves. An undescribed species of Alseuosmia from the Hooker (1852-5, 1864), with the benefit of Waimarino region of the Volcanic Plateau has been additional Colenso and Sinclair material, reduced known for some time (Cockayne 1928, p. 179; A. P. Cunningham's species to four, but stated that these Druce in Atkinson 1971). The following description four species were "excessively variable". -

State of New York City's Plants 2018
STATE OF NEW YORK CITY’S PLANTS 2018 Daniel Atha & Brian Boom © 2018 The New York Botanical Garden All rights reserved ISBN 978-0-89327-955-4 Center for Conservation Strategy The New York Botanical Garden 2900 Southern Boulevard Bronx, NY 10458 All photos NYBG staff Citation: Atha, D. and B. Boom. 2018. State of New York City’s Plants 2018. Center for Conservation Strategy. The New York Botanical Garden, Bronx, NY. 132 pp. STATE OF NEW YORK CITY’S PLANTS 2018 4 EXECUTIVE SUMMARY 6 INTRODUCTION 10 DOCUMENTING THE CITY’S PLANTS 10 The Flora of New York City 11 Rare Species 14 Focus on Specific Area 16 Botanical Spectacle: Summer Snow 18 CITIZEN SCIENCE 20 THREATS TO THE CITY’S PLANTS 24 NEW YORK STATE PROHIBITED AND REGULATED INVASIVE SPECIES FOUND IN NEW YORK CITY 26 LOOKING AHEAD 27 CONTRIBUTORS AND ACKNOWLEGMENTS 30 LITERATURE CITED 31 APPENDIX Checklist of the Spontaneous Vascular Plants of New York City 32 Ferns and Fern Allies 35 Gymnosperms 36 Nymphaeales and Magnoliids 37 Monocots 67 Dicots 3 EXECUTIVE SUMMARY This report, State of New York City’s Plants 2018, is the first rankings of rare, threatened, endangered, and extinct species of what is envisioned by the Center for Conservation Strategy known from New York City, and based on this compilation of The New York Botanical Garden as annual updates thirteen percent of the City’s flora is imperiled or extinct in New summarizing the status of the spontaneous plant species of the York City. five boroughs of New York City. This year’s report deals with the City’s vascular plants (ferns and fern allies, gymnosperms, We have begun the process of assessing conservation status and flowering plants), but in the future it is planned to phase in at the local level for all species. -

Plastid Phylogenomic Insights Into the Evolution of the Caprifoliaceae S.L. (Dipsacales)
Molecular Phylogenetics and Evolution 142 (2020) 106641 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Plastid phylogenomic insights into the evolution of the Caprifoliaceae s.l. T (Dipsacales) Hong-Xin Wanga,1, Huan Liub,c,1, Michael J. Moored, Sven Landreine, Bing Liuf,g, Zhi-Xin Zhua, ⁎ Hua-Feng Wanga, a Key Laboratory of Tropical Biological Resources of Ministry of Education, School of Life and Pharmaceutical Sciences, Hainan University, Haikou 570228, China b BGI-Shenzhen, Beishan Industrial Zone, Yantian District, Shenzhen 518083, China c State Key Laboratory of Agricultural Genomics, BGI-Shenzhen, Shenzhen 518083, China d Department of Biology, Oberlin College, Oberlin, OH 44074, USA e Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun, 666303, China f State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Science, Beijing 100093, China g Sino-African Joint Research Centre, Chinese Academy of Science, Wuhan 430074, China ARTICLE INFO ABSTRACT Keywords: The family Caprifoliaceae s.l. is an asterid angiosperm clade of ca. 960 species, most of which are distributed in Caprifoliaceae s.l. temperate regions of the northern hemisphere. Recent studies show that the family comprises seven major Dipsacales clades: Linnaeoideae, Zabelia, Morinoideae, Dipsacoideae, Valerianoideae, Caprifolioideae, and Diervilloideae. Plastome However, its phylogeny at the subfamily or genus level remains controversial, and the backbone relationships Phylogenetics among subfamilies are incompletely resolved. In this study, we utilized complete plastome sequencing to resolve the relationships among the subfamilies of the Caprifoliaceae s.l. and clarify several long-standing controversies. We generated and analyzed plastomes of 48 accessions of Caprifoliaceae s.l., representing 44 species, six sub- families and one genus. -

Bio 308-Course Guide
COURSE GUIDE BIO 308 BIOGEOGRAPHY Course Team Dr. Kelechi L. Njoku (Course Developer/Writer) Professor A. Adebanjo (Programme Leader)- NOUN Abiodun E. Adams (Course Coordinator)-NOUN NATIONAL OPEN UNIVERSITY OF NIGERIA BIO 308 COURSE GUIDE National Open University of Nigeria Headquarters 14/16 Ahmadu Bello Way Victoria Island Lagos Abuja Office No. 5 Dar es Salaam Street Off Aminu Kano Crescent Wuse II, Abuja e-mail: [email protected] URL: www.nou.edu.ng Published by National Open University of Nigeria Printed 2013 ISBN: 978-058-434-X All Rights Reserved Printed by: ii BIO 308 COURSE GUIDE CONTENTS PAGE Introduction ……………………………………......................... iv What you will Learn from this Course …………………............ iv Course Aims ……………………………………………............ iv Course Objectives …………………………………………....... iv Working through this Course …………………………….......... v Course Materials ………………………………………….......... v Study Units ………………………………………………......... v Textbooks and References ………………………………........... vi Assessment ……………………………………………….......... vi End of Course Examination and Grading..................................... vi Course Marking Scheme................................................................ vii Presentation Schedule.................................................................... vii Tutor-Marked Assignment ……………………………….......... vii Tutors and Tutorials....................................................................... viii iii BIO 308 COURSE GUIDE INTRODUCTION BIO 308: Biogeography is a one-semester, 2 credit- hour course in Biology. It is a 300 level, second semester undergraduate course offered to students admitted in the School of Science and Technology, School of Education who are offering Biology or related programmes. The course guide tells you briefly what the course is all about, what course materials you will be using and how you can work your way through these materials. It gives you some guidance on your Tutor- Marked Assignments. There are Self-Assessment Exercises within the body of a unit and/or at the end of each unit. -

The Origin of the Bifurcating Style in Asteraceae (Compositae)
Annals of Botany 117: 1009–1021, 2016 doi:10.1093/aob/mcw033, available online at www.aob.oxfordjournals.org The origin of the bifurcating style in Asteraceae (Compositae) Liliana Katinas1,2,*, Marcelo P. Hernandez 2, Ana M. Arambarri2 and Vicki A. Funk3 1Division Plantas Vasculares, Museo de La Plata, La Plata, Argentina, 2Laboratorio de Morfologıa Comparada de Espermatofitas (LAMCE), Facultad de Ciencias Agrarias y Forestales, Universidad Nacional de La Plata, La Plata, Argentina and 3Department of Botany, NMNH, Smithsonian Institution, Washington D.C., USA *For correspondence. E-mail [email protected] Received: 20 November 2015 Returned for revision: 22 December 2015 Accepted: 8 January 2016 Published electronically: 20 April 2016 Background and Aims The plant family Asteraceae (Compositae) exhibits remarkable morphological variation in the styles of its members. Lack of studies on the styles of the sister families to Asteraceae, Goodeniaceae and Calyceraceae, obscures our understanding of the origin and evolution of this reproductive feature in these groups. The aim of this work was to perform a comparative study of style morphology and to discuss the relevance of im- portant features in the evolution of Asteraceae and its sister families. Methods The histochemistry, venation and general morphology of the styles of members of Goodeniaceae, Calyceraceae and early branching lineages of Asteraceae were analysed and put in a phylogenetic framework to dis- cuss the relevance of style features in the evolution of these families. Key Results The location of lipophilic substances allowed differentiation of receptive from non-receptive style papillae, and the style venation in Goodeniaceae and Calyceraceae proved to be distinctive. -

Checklist of the Vascular Plants of Redwood National Park
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 9-17-2018 Checklist of the Vascular Plants of Redwood National Park James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Checklist of the Vascular Plants of Redwood National Park" (2018). Botanical Studies. 85. https://digitalcommons.humboldt.edu/botany_jps/85 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. A CHECKLIST OF THE VASCULAR PLANTS OF THE REDWOOD NATIONAL & STATE PARKS James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State Univerity Arcata, California 14 September 2018 The Redwood National and State Parks are located in Del Norte and Humboldt counties in coastal northwestern California. The national park was F E R N S established in 1968. In 1994, a cooperative agreement with the California Department of Parks and Recreation added Del Norte Coast, Prairie Creek, Athyriaceae – Lady Fern Family and Jedediah Smith Redwoods state parks to form a single administrative Athyrium filix-femina var. cyclosporum • northwestern lady fern unit. Together they comprise about 133,000 acres (540 km2), including 37 miles of coast line. Almost half of the remaining old growth redwood forests Blechnaceae – Deer Fern Family are protected in these four parks. -

Phylogeny and Phylogenetic Taxonomy of Dipsacales, with Special Reference to Sinadoxa and Tetradoxa (Adoxaceae)
PHYLOGENY AND PHYLOGENETIC TAXONOMY OF DIPSACALES, WITH SPECIAL REFERENCE TO SINADOXA AND TETRADOXA (ADOXACEAE) MICHAEL J. DONOGHUE,1 TORSTEN ERIKSSON,2 PATRICK A. REEVES,3 AND RICHARD G. OLMSTEAD 3 Abstract. To further clarify phylogenetic relationships within Dipsacales,we analyzed new and previously pub- lished rbcL sequences, alone and in combination with morphological data. We also examined relationships within Adoxaceae using rbcL and nuclear ribosomal internal transcribed spacer (ITS) sequences. We conclude from these analyses that Dipsacales comprise two major lineages:Adoxaceae and Caprifoliaceae (sensu Judd et al.,1994), which both contain elements of traditional Caprifoliaceae.Within Adoxaceae, the following relation- ships are strongly supported: (Viburnum (Sambucus (Sinadoxa (Tetradoxa, Adoxa)))). Combined analyses of C ap ri foliaceae yield the fo l l ow i n g : ( C ap ri folieae (Diervilleae (Linnaeeae (Morinaceae (Dipsacaceae (Triplostegia,Valerianaceae)))))). On the basis of these results we provide phylogenetic definitions for the names of several major clades. Within Adoxaceae, Adoxina refers to the clade including Sinadoxa, Tetradoxa, and Adoxa.This lineage is marked by herbaceous habit, reduction in the number of perianth parts,nectaries of mul- ticellular hairs on the perianth,and bifid stamens. The clade including Morinaceae,Valerianaceae, Triplostegia, and Dipsacaceae is here named Valerina. Probable synapomorphies include herbaceousness,presence of an epi- calyx (lost or modified in Valerianaceae), reduced endosperm,and distinctive chemistry, including production of monoterpenoids. The clade containing Valerina plus Linnaeeae we name Linnina. This lineage is distinguished by reduction to four (or fewer) stamens, by abortion of two of the three carpels,and possibly by supernumerary inflorescences bracts. Keywords: Adoxaceae, Caprifoliaceae, Dipsacales, ITS, morphological characters, phylogeny, phylogenetic taxonomy, phylogenetic nomenclature, rbcL, Sinadoxa, Tetradoxa. -

Part I Chinese Plant Names Index 2010-2017
This Book is Sponsored by Shanghai Chenshan Botanical Garden 上海辰山植物园 Shanghai Chenshan Plant Science Research Center, Chinese Academy of Sciences 中国科学院上海辰山植物科学研究中心 Special Fund for Scientific Research of Shanghai Landscaping & City Appearance Administrative Bureau (G182415) 上海市绿化和市容管理局科研专项 (G182415) National Specimen Information Infrastructure, 2018 Special Funds 中国国家标本平台 2018 年度专项 Shanghai Sailing Program (14YF1413800) 上海市青年科技英才扬帆计划 (14YF1413800) Chinese Plant Names Index 2010-2017 DU Cheng & MA Jin-shuang Chinese Plant Names Index 2010-2017 中国植物名称索引 2010-2017 DU Cheng & MA Jin-shuang Abstract The first two volumes of Chinese Plant Names Index (CPNI) cover the years 2000 through 2009, with entries 1 through 5,516, and 2010 through 2017, with entries 5,517 through 10,795. A unique entry is generated for the specific name of each taxon in a specific publication. Taxonomic treatments cover all novelties at the rank of family, genus, species, subspecies, variety, form and named hybrid taxa, new name changes (new combinations and new names), new records, new synonyms and new typifications for vascular plants reported or recorded from China. Detailed information on the place of publication, including author, publication name, year of publication, volume, issue, and page number, are given in detail. Type specimens and collects information for the taxa and their distribution in China, as well as worldwide, are also provided. The bibliographies were compiled from 182 journals and 138 monographs or books published worldwide. In addition, more than 400 herbaria preserve type specimens of Chinese plants are also listed as an appendix. This book can be used as a basic material for Chinese vascular plant taxonomy, and as a reference for researchers in biodiversity research, environmental protection, forestry and medicinal botany. -

Anatomy and Affinities of Penthorum Melanie Lynn Haskins
University of Richmond UR Scholarship Repository Biology Faculty Publications Biology 2-1987 Anatomy and Affinities of Penthorum Melanie Lynn Haskins W. John Hayden University of Richmond, [email protected] Follow this and additional works at: http://scholarship.richmond.edu/biology-faculty-publications Part of the Botany Commons, Other Plant Sciences Commons, and the Plant Biology Commons Recommended Citation Haskins, Melanie Lynn, and W. John Hayden. "Anatomy and Affinities of Penthorum." American Journal of Botany 74, no. 2 (February 1987): 164-77. This Article is brought to you for free and open access by the Biology at UR Scholarship Repository. It has been accepted for inclusion in Biology Faculty Publications by an authorized administrator of UR Scholarship Repository. For more information, please contact [email protected]. Amer. J. Bot. 74(2): 164-177. 1987. ANATOMY AND AFFINITIES OF PENTHORUM' MELANIE L. HASKINS AND W. JOHN HAYDEN Department of Biology, University of Richmond, Richmond, Virginia 23173 ABSTRACT The genus Penthorum L. consists of two species of perennial herbs, P. sedoides of eastern North America and P. chinense ofeastern Asia. Pentho rum has long been considered intermediate between Crassulaceae and Saxifragaceae. An anatomical study of both species was undertaken to contribute to a better understanding of the relationships ofthese plants. Prominent anatomical features of Penthorum include: an aerenchymatous cortex and closely-spaced collateral vascular bundles of stems; one-trace unilacunar nodes; brochidodromous venation, rosoid teeth bearing hydathodes, and anomocytic stomata of leaves; angular vessel elements with many-barred scalariform perforation plates and alternate to scattered intervascular pits; thin-walled non septate fiber-tracheids; abundant homocellular erect uniseriate and biseriate rays; and absence of axial xylem parenchyma. -
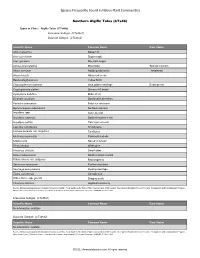
Algific Talus (Cts46)
Species Frequently Found in Native Plant Communities Southern Algific Talus (CTs46) Types in Class: Algific Talus (CTs46a) Limestone Subtype (CTs46a1) Dolomite Subtype (CTs46a2) Scientific Name Column1 Common Name Rare Status Abies balsamea Balsam fir Acer saccharum Sugar maple Acer spicatum Mountain maple Adoxa moschatellina Moschatel Special Concern Allium cernuum Nodding wild onion Threatened Arabis hirsuta Hairy rock cress Betula alleghaniensis Yellow birch Chrysosplenium iowense Iowa golden saxifrage Endangered Cryptogramma stelleri Slender cliff brake Cystopteris bulbifera Bulblet fern Dicentra cucullaria Dutchman's breeches Enemion biternatum False rue anemone Gymnocarpium robertianum Northern oak fern Impatiens spp. touch-me-not Impatiens capensis Spotted touch-me-not Impatiens pallida Pale touch-me-not Laportea canadensis Wood nettle Linnaea borealis var. longiflora Twinflower Mertensia paniculata Panicled bluebells Mitella nuda Naked miterwort Pinus strobus White pine Rhamnus alnifolia Dwarf alder Ribes hudsonianum Northern black currant Rubus idaeus var. strigosus Red raspberry Sambucus racemosa Red-berried elder Saxifraga pensylvanica Swamp saxifrage Taxus canadensis Canada yew Urtica dioica ssp. gracilis Stinging nettle Viburnum trilobum Highbush cranberry Source: Minnesota Department of Natural Resources (2005). Field Guide to the Native Plant Communities of Minnesota: The Eastern Broadleaf Forest Province. Ecological Land Classification Program, Minnesota County Biological Survey, and Natural Heritage and Nongame Research Program. MNDNR St. Paul, MN. Limestone Subtype (CTs46a1) Scientific Name Column1 Common Name Rare Status No information available Dolomite Subtype (CTs46a2) Scientific Name Column1 Common Name Rare Status No information available Source: Minnesota Department of Natural Resources (2005). Field Guide to the Native Plant Communities of Minnesota: The Eastern Broadleaf Forest Province. Ecological Land Classification Program, Minnesota County Biological Survey, and Natural Heritage and Nongame Research Program.