Molecular Basis of Development in Petaloid Monocot Flowers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Well-Known Plants in Each Angiosperm Order
Well-known plants in each angiosperm order This list is generally from least evolved (most ancient) to most evolved (most modern). (I’m not sure if this applies for Eudicots; I’m listing them in the same order as APG II.) The first few plants are mostly primitive pond and aquarium plants. Next is Illicium (anise tree) from Austrobaileyales, then the magnoliids (Canellales thru Piperales), then monocots (Acorales through Zingiberales), and finally eudicots (Buxales through Dipsacales). The plants before the eudicots in this list are considered basal angiosperms. This list focuses only on angiosperms and does not look at earlier plants such as mosses, ferns, and conifers. Basal angiosperms – mostly aquatic plants Unplaced in order, placed in Amborellaceae family • Amborella trichopoda – one of the most ancient flowering plants Unplaced in order, placed in Nymphaeaceae family • Water lily • Cabomba (fanwort) • Brasenia (watershield) Ceratophyllales • Hornwort Austrobaileyales • Illicium (anise tree, star anise) Basal angiosperms - magnoliids Canellales • Drimys (winter's bark) • Tasmanian pepper Laurales • Bay laurel • Cinnamon • Avocado • Sassafras • Camphor tree • Calycanthus (sweetshrub, spicebush) • Lindera (spicebush, Benjamin bush) Magnoliales • Custard-apple • Pawpaw • guanábana (soursop) • Sugar-apple or sweetsop • Cherimoya • Magnolia • Tuliptree • Michelia • Nutmeg • Clove Piperales • Black pepper • Kava • Lizard’s tail • Aristolochia (birthwort, pipevine, Dutchman's pipe) • Asarum (wild ginger) Basal angiosperms - monocots Acorales -

Full of Beans: a Study on the Alignment of Two Flowering Plants Classification Systems
Full of beans: a study on the alignment of two flowering plants classification systems Yi-Yun Cheng and Bertram Ludäscher School of Information Sciences, University of Illinois at Urbana-Champaign, USA {yiyunyc2,ludaesch}@illinois.edu Abstract. Advancements in technologies such as DNA analysis have given rise to new ways in organizing organisms in biodiversity classification systems. In this paper, we examine the feasibility of aligning two classification systems for flowering plants using a logic-based, Region Connection Calculus (RCC-5) ap- proach. The older “Cronquist system” (1981) classifies plants using their mor- phological features, while the more recent Angiosperm Phylogeny Group IV (APG IV) (2016) system classifies based on many new methods including ge- nome-level analysis. In our approach, we align pairwise concepts X and Y from two taxonomies using five basic set relations: congruence (X=Y), inclusion (X>Y), inverse inclusion (X<Y), overlap (X><Y), and disjointness (X!Y). With some of the RCC-5 relationships among the Fabaceae family (beans family) and the Sapindaceae family (maple family) uncertain, we anticipate that the merging of the two classification systems will lead to numerous merged solutions, so- called possible worlds. Our research demonstrates how logic-based alignment with ambiguities can lead to multiple merged solutions, which would not have been feasible when aligning taxonomies, classifications, or other knowledge or- ganization systems (KOS) manually. We believe that this work can introduce a novel approach for aligning KOS, where merged possible worlds can serve as a minimum viable product for engaging domain experts in the loop. Keywords: taxonomy alignment, KOS alignment, interoperability 1 Introduction With the advent of large-scale technologies and datasets, it has become increasingly difficult to organize information using a stable unitary classification scheme over time. -

Reconstructing the Basal Angiosperm Phylogeny: Evaluating Information Content of Mitochondrial Genes
55 (4) • November 2006: 837–856 Qiu & al. • Basal angiosperm phylogeny Reconstructing the basal angiosperm phylogeny: evaluating information content of mitochondrial genes Yin-Long Qiu1, Libo Li, Tory A. Hendry, Ruiqi Li, David W. Taylor, Michael J. Issa, Alexander J. Ronen, Mona L. Vekaria & Adam M. White 1Department of Ecology & Evolutionary Biology, The University Herbarium, University of Michigan, Ann Arbor, Michigan 48109-1048, U.S.A. [email protected] (author for correspondence). Three mitochondrial (atp1, matR, nad5), four chloroplast (atpB, matK, rbcL, rpoC2), and one nuclear (18S) genes from 162 seed plants, representing all major lineages of gymnosperms and angiosperms, were analyzed together in a supermatrix or in various partitions using likelihood and parsimony methods. The results show that Amborella + Nymphaeales together constitute the first diverging lineage of angiosperms, and that the topology of Amborella alone being sister to all other angiosperms likely represents a local long branch attrac- tion artifact. The monophyly of magnoliids, as well as sister relationships between Magnoliales and Laurales, and between Canellales and Piperales, are all strongly supported. The sister relationship to eudicots of Ceratophyllum is not strongly supported by this study; instead a placement of the genus with Chloranthaceae receives moderate support in the mitochondrial gene analyses. Relationships among magnoliids, monocots, and eudicots remain unresolved. Direct comparisons of analytic results from several data partitions with or without RNA editing sites show that in multigene analyses, RNA editing has no effect on well supported rela- tionships, but minor effect on weakly supported ones. Finally, comparisons of results from separate analyses of mitochondrial and chloroplast genes demonstrate that mitochondrial genes, with overall slower rates of sub- stitution than chloroplast genes, are informative phylogenetic markers, and are particularly suitable for resolv- ing deep relationships. -

Bilirubin: an Animal Pigment in the Zingiberales and Diverse Angiosperm Orders Cary L
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 11-5-2010 Bilirubin: an Animal Pigment in the Zingiberales and Diverse Angiosperm Orders Cary L. Pirone Florida International University, [email protected] DOI: 10.25148/etd.FI10122201 Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Biochemistry Commons, and the Botany Commons Recommended Citation Pirone, Cary L., "Bilirubin: an Animal Pigment in the Zingiberales and Diverse Angiosperm Orders" (2010). FIU Electronic Theses and Dissertations. 336. https://digitalcommons.fiu.edu/etd/336 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida BILIRUBIN: AN ANIMAL PIGMENT IN THE ZINGIBERALES AND DIVERSE ANGIOSPERM ORDERS A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in BIOLOGY by Cary Lunsford Pirone 2010 To: Dean Kenneth G. Furton College of Arts and Sciences This dissertation, written by Cary Lunsford Pirone, and entitled Bilirubin: An Animal Pigment in the Zingiberales and Diverse Angiosperm Orders, having been approved in respect to style and intellectual content, is referred to you for judgment. We have read this dissertation and recommend that it be approved. ______________________________________ Bradley C. Bennett ______________________________________ Timothy M. Collins ______________________________________ Maureen A. Donnelly ______________________________________ John. T. Landrum ______________________________________ J. Martin Quirke ______________________________________ David W. Lee, Major Professor Date of Defense: November 5, 2010 The dissertation of Cary Lunsford Pirone is approved. -

ABSTRACTS 117 Systematics Section, BSA / ASPT / IOPB
Systematics Section, BSA / ASPT / IOPB 466 HARDY, CHRISTOPHER R.1,2*, JERROLD I DAVIS1, breeding system. This effectively reproductively isolates the species. ROBERT B. FADEN3, AND DENNIS W. STEVENSON1,2 Previous studies have provided extensive genetic, phylogenetic and 1Bailey Hortorium, Cornell University, Ithaca, NY 14853; 2New York natural selection data which allow for a rare opportunity to now Botanical Garden, Bronx, NY 10458; 3Dept. of Botany, National study and interpret ontogenetic changes as sources of evolutionary Museum of Natural History, Smithsonian Institution, Washington, novelties in floral form. Three populations of M. cardinalis and four DC 20560 populations of M. lewisii (representing both described races) were studied from initiation of floral apex to anthesis using SEM and light Phylogenetics of Cochliostema, Geogenanthus, and microscopy. Allometric analyses were conducted on data derived an undescribed genus (Commelinaceae) using from floral organs. Sympatric populations of the species from morphology and DNA sequence data from 26S, 5S- Yosemite National Park were compared. Calyces of M. lewisii initi- NTS, rbcL, and trnL-F loci ate later than those of M. cardinalis relative to the inner whorls, and sepals are taller and more acute. Relative times of initiation of phylogenetic study was conducted on a group of three small petals, sepals and pistil are similar in both species. Petal shapes dif- genera of neotropical Commelinaceae that exhibit a variety fer between species throughout development. Corolla aperture of unusual floral morphologies and habits. Morphological A shape becomes dorso-ventrally narrow during development of M. characters and DNA sequence data from plastid (rbcL, trnL-F) and lewisii, and laterally narrow in M. -

Angiosperm Clades in the Potomac Group: What Have We Learned Since 1977?
UC Davis UC Davis Previously Published Works Title Angiosperm clades in the potomac group: What have we learned since 1977? Permalink https://escholarship.org/uc/item/5z89c18z Journal Bulletin of the Peabody Museum of Natural History, 55(2-3) ISSN 0079-032X Authors Doyle, JA Upchurch, GR Publication Date 2014-10-01 DOI 10.3374/014.055.0203 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Angiosperm Clades in the Potomac Group: What Have We Learned since 1977? James A. Doyle1 and Garland R. Upchurch, Jr.2 1 Corresponding author: Department of Evolution and Ecology, University of California, Davis CA 95616 USA —email: [email protected] 2 Department of Biology, Texas State University, San Marcos TX 78666 USA —email: [email protected] ABSTRACT In their 1977 study on Potomac Group angiosperms, Hickey and Doyle made only broad com- parisons with living taxa. Newer data, especially discoveries of fossil flowers in the Potomac and coeval deposits and increasingly robust molecular phylogenies of living angiosperms, allow more precise phylogenetic placement of fossils. Hickey and Doyle compared most early Potomac leaves (Aptian–early Albian) with “magnoliids,” a paraphyletic group as then defined, but several clades can now be recognized. Leaves and dispersed cuticles share epidermal features with woody mem- bers of the basal ANITA grade, and in some cases crown group Austrobaileyales, whose presence is confirmed by flowers called Anacostia. Aptian–Albian flowers (Monetianthus, Carpestella) and whole plants (Pluricarpellatia) are nested in crown group Nymphaeales; Potomac reniform leaves could belong to this clade. Several Potomac leaves have chloranthoid teeth, venation, and oppo- site phyllotaxis consistent with Chloranthaceae, while Aptian to Cenomanian flowers reveal the presence of both crown group Chloranthaceae (Asteropollis plant, near Hedyosmum) and stem relatives of this family and/or Ceratophyllum (Canrightia, Zlatkocarpus, Pennipollis plant, possi- bly Appomattoxia). -

Eudicots Monocots Stems Embryos Roots Leaf Venation Pollen Flowers
Monocots Eudicots Embryos One cotyledon Two cotyledons Leaf venation Veins Veins usually parallel usually netlike Stems Vascular tissue Vascular tissue scattered usually arranged in ring Roots Root system usually Taproot (main root) fibrous (no main root) usually present Pollen Pollen grain with Pollen grain with one opening three openings Flowers Floral organs usually Floral organs usually in in multiples of three multiples of four or five © 2014 Pearson Education, Inc. 1 Reproductive shoot (flower) Apical bud Node Internode Apical bud Shoot Vegetative shoot system Blade Leaf Petiole Axillary bud Stem Taproot Lateral Root (branch) system roots © 2014 Pearson Education, Inc. 2 © 2014 Pearson Education, Inc. 3 Storage roots Pneumatophores “Strangling” aerial roots © 2014 Pearson Education, Inc. 4 Stolon Rhizome Root Rhizomes Stolons Tubers © 2014 Pearson Education, Inc. 5 Spines Tendrils Storage leaves Stem Reproductive leaves Storage leaves © 2014 Pearson Education, Inc. 6 Dermal tissue Ground tissue Vascular tissue © 2014 Pearson Education, Inc. 7 Parenchyma cells with chloroplasts (in Elodea leaf) 60 µm (LM) © 2014 Pearson Education, Inc. 8 Collenchyma cells (in Helianthus stem) (LM) 5 µm © 2014 Pearson Education, Inc. 9 5 µm Sclereid cells (in pear) (LM) 25 µm Cell wall Fiber cells (cross section from ash tree) (LM) © 2014 Pearson Education, Inc. 10 Vessel Tracheids 100 µm Pits Tracheids and vessels (colorized SEM) Perforation plate Vessel element Vessel elements, with perforated end walls Tracheids © 2014 Pearson Education, Inc. 11 Sieve-tube elements: 3 µm longitudinal view (LM) Sieve plate Sieve-tube element (left) and companion cell: Companion cross section (TEM) cells Sieve-tube elements Plasmodesma Sieve plate 30 µm Nucleus of companion cell 15 µm Sieve-tube elements: longitudinal view Sieve plate with pores (LM) © 2014 Pearson Education, Inc. -
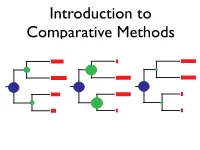
BM CC EB What Can We Learn from a Tree?
Introduction to Comparative Methods BM CC EB What can we learn from a tree? Net diversification (r) Relative extinction (ε) Peridiscaceae Peridiscaceae yllaceae yllaceae h h atop atop Proteaceae Proteaceae r r Ce Ce Tr oc T ho r M M o de c y y H H C C h r r e e a D D a o o nd o e e G G a a m m t t a a d r P r P h h e e u u c c p A p A r e a a e e a a a c a c n n a i B i B h h n d m d l m a l a m m e a e a e e t t n n c u u n n i i d i i e e e e o n o n p n p n a e a S e e S e e n n x x i i r c c a a n o n o p p h g e h g ae e l r a l r a a a a a a i i a a a e a e i b i b h y d c h d c i y i c a a c x c x c c G I a G I a n c n c c c y l y l t a a t a a e e e e e i l c i l c m l m l e c e c f a e a a f a e a a l r r l c c a i i r l e t e t a a r l a a e e u u u u o a o a a a c a c a a l a l e e e b b a a a a e e c e e c a a s c s c c e l c e l e e g e g e a a a a e e n n s e e s e e e e a a a P a P e e N N u u u S u S a e a e a a e e c c l n a l n e e a e e a e a e a e a e r a r a c c C i C i R R a e a e a e a e r c r c A A a d a a d a e i e i phanopetalaceae s r e ph s r e a a s e c s e c e e u u b a a b a e e P P r r l l e n e a a a a m m entho e e e Ha a H o a c r e c r e nt B B e p e e e c e e c a c c h e a p a a p a lo lo l l a a e s o t e s i a r a i a r r r r r a n e a n e a b a l b t a t gaceae e g e ceae a c a s c s a z e z M i a e M i a c a d e a d e ae e ae r e r a e a e a a a c c ce r e r L L i i ac a Vitaceae Vi r r C C e e ta v e v e a a c a a e ea p e ap c a c a e a P P e e l l e Ge G e e ae a t t e e p p r r ce c an u an -

Field Identification of the 50 Most Common Plant Families in Temperate Regions
Field identification of the 50 most common plant families in temperate regions (including agricultural, horticultural, and wild species) by Lena Struwe [email protected] © 2016, All rights reserved. Note: Listed characteristics are the most common characteristics; there might be exceptions in rare or tropical species. This compendium is available for free download without cost for non- commercial uses at http://www.rci.rutgers.edu/~struwe/. The author welcomes updates and corrections. 1 Overall phylogeny – living land plants Bryophytes Mosses, liverworts, hornworts Lycophytes Clubmosses, etc. Ferns and Fern Allies Ferns, horsetails, moonworts, etc. Gymnosperms Conifers, pines, cycads and cedars, etc. Magnoliids Monocots Fabids Ranunculales Rosids Malvids Caryophyllales Ericales Lamiids The treatment for flowering plants follows the APG IV (2016) Campanulids classification. Not all branches are shown. © Lena Struwe 2016, All rights reserved. 2 Included families (alphabetical list): Amaranthaceae Geraniaceae Amaryllidaceae Iridaceae Anacardiaceae Juglandaceae Apiaceae Juncaceae Apocynaceae Lamiaceae Araceae Lauraceae Araliaceae Liliaceae Asphodelaceae Magnoliaceae Asteraceae Malvaceae Betulaceae Moraceae Boraginaceae Myrtaceae Brassicaceae Oleaceae Bromeliaceae Orchidaceae Cactaceae Orobanchaceae Campanulaceae Pinaceae Caprifoliaceae Plantaginaceae Caryophyllaceae Poaceae Convolvulaceae Polygonaceae Cucurbitaceae Ranunculaceae Cupressaceae Rosaceae Cyperaceae Rubiaceae Equisetaceae Rutaceae Ericaceae Salicaceae Euphorbiaceae Scrophulariaceae -

Basal Eudicots
9/12/2019 Basal Eudicots Dicots in transition between Basal Grade (Dicot) Angiosperms & Core Eudicots • Many stamens and, usu, carpels • Perianth reduced in number, though still poorly differentiated • Parts generally free (some with fusion in G) • Triaperturate pollen. Tricolpate pollen from the Lamiales (USDA Pollen Lab) Ranunculaceae (buttercup family) Herbs to vines or woody vines Lvs simple, compound, to dissected, all with sheathing bases & membranous margins. Perianth whorls poorly differentiated Ca4-5-many Co4-5-many Amany Gseveral-many on convex receptacle 1 9/12/2019 Ranunculaceae (buttercup family) Ranunculus (UMass Extension) (PM Dziuk, 2006) (UMass Extension) Ranunculaceae (buttercup family) Ranunculus 2 9/12/2019 Ranunculaceae (buttercup family) USDA Plants Database Clematis Papaveraceae (poppy family) Herbs (shrubs) often with milky or colored latex. Lvs simple to pinnate to pinnately much dissected with or without sheathing base with membranous margins. Perianth whorls well differentiated between 2 caducous sepals and 4‐8 showy petals. Ca2,caducous Co4-8 Amany or 6 G[2-many],short to absent style 3 9/12/2019 Papaveraceae (poppy family) Papaver orientale Papaveraceae (poppy family) Papaver somniferum 4 9/12/2019 Papaveraceae (poppy family) Papaver somniferum Papaveraceae (poppy family) Chelidonium (celandine‐poppy) 5 9/12/2019 Papaveraceae (poppy family) Sanguinaria http://alivestructures.blogspot.com/ Dicentra 6 9/12/2019 Dicentra Papaveraceae (poppy family) Lamprocapnos http://alivestructures.blogspot.com/ 7 9/12/2019 -

Djvu Document
Cenomanian Angiosperm Leaf Megafossils, Dakota Formation, Rose Creek Locality, Jefferson County, Southeastern Nebraska By GARLAND R. UPCHURCH, JR. and DAVID L. DILCHER U.S. GEOLOGICAL SURVEY BULLETIN 1915 DEPARTMENT OF THE INTERIOR MANUEL LUJAN, JR., Secretary U.S. GEOLOGICAL SURVEY Dallas L. Peck, Director Any use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the U.S. Government. UNITED STATES GOVERNMENT PRINTING OFFICE: 1990 For sale by the Books and Open-File Reports Section U.S. Geological Survey Federal Center Box 25425 Denver. CO 80225 Library of Congress Cataloging-in-Publication Data Upchurch, Garland R. Cenomanian angiosperm leaf megafossils, Dakota Formation, Rose Creek locality, Jefferson County, southeastern Nebraska / by Garland R. Upchurch, Jr., and David L. Dilcher. p. cm.-(U.S. Geological Survey bulletin; 1915) Includes bibliographical references. Supt. of Docs. no.: 1 19.3:1915. 1. Leaves, Fossil-Nebraska-Jefferson County. 2. Paleobotany-Cretaceous. 3. Paleobotany-Nebraska-Jefferson County. I. Dilcher, David L. II. Title. III. Series. QE75.B9 no. 1915 [QE983] 557.3 s-dc20 90-2855 [561'.2]CIP CONTENTS Abstract 1 Introduction 1 Acknowledgments 2 Materials and methods 2 Criteria for classification 3 Geological setting and description of fossil plant locality 4 Floristic composition 7 Evolutionary considerations 8 Ecological considerations 9 Key to leaf types at Rose Creek 10 Systematics 12 Magnoliales 12 Laurales 13 cf. Illiciales 30 Magnoliidae order -

Fossil Calibration of Magnoliidae, an Ancient Lineage of Angiosperms
Palaeontologia Electronica palaeo-electronica.org Fossil calibration of Magnoliidae, an ancient lineage of angiosperms Julien Massoni, James Doyle, and Hervé Sauquet ABSTRACT In order to investigate the diversification of angiosperms, an accurate temporal framework is needed. Molecular dating methods thoroughly calibrated with the fossil record can provide estimates of this evolutionary time scale. Because of their position in the phylogenetic tree of angiosperms, Magnoliidae (10,000 species) are of primary importance for the investigation of the evolutionary history of flowering plants. The rich fossil record of the group, beginning in the Cretaceous, has a global distribution. Among the hundred extinct species of Magnoliidae described, several have been included in phylogenetic analyses alongside extant species, providing reliable calibra- tion points for molecular dating studies. Until now, few fossils have been used as cali- bration points of Magnoliidae, and detailed justifications of their phylogenetic position and absolute age have been lacking. Here, we review the position and ages for 10 fos- sils of Magnoliidae, selected because of their previous inclusion in phylogenetic analy- ses of extant and fossil taxa. This study allows us to propose an updated calibration scheme for dating the evolutionary history of Magnoliidae. Julien Massoni. Laboratoire Ecologie, Systématique, Evolution, Université Paris-Sud, CNRS UMR 8079, 91405 Orsay, France. [email protected] James Doyle. Department of Evolution and Ecology, University of California, Davis, CA 95616, USA. [email protected] Hervé Sauquet. Laboratoire Ecologie, Systématique, Evolution, Université Paris-Sud, CNRS UMR 8079, 91405 Orsay, France. [email protected] Keywords: fossil calibration; Canellales; Laurales; Magnoliales; Magnoliidae; Piperales PE Article Number: 18.1.2FC Copyright: Palaeontological Association February 2015 Submission: 10 October 2013.