Structure of a Bacterial Enzyme Regulated by Phosphorylation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dehydrogenase Gene of Entamoeba Histolytica
Proc. Nad. Acad. Sci. USA Vol. 89, pp. 10188-10192, November 1992 Medical Sciences Cloning and expression of an NADP+-dependent alcohol dehydrogenase gene of Entamoeba histolytica (amebiasis/protozoan parasite/fermentation/inhibitors) AJIT KUMAR*, PEI-SHEN SHENtt, STEVEN DESCOTEAUX*, JAN POHL§, GORDON BAILEYt, AND JOHN SAMUELSON*1II *Department of Tropical Public Health, Harvard School of Public Health, Boston, MA 02115; tDepartment of Biochemistry, Morehouse School of Medicine, Atlanta, GA 30310; §Microchemical Facility, Emory University, Atlanta, GA 30322; and 'Department of Pathology, New England Deaconess Hospital, Boston, MA 02215 Communicated by Elkan Blout, June 29, 1992 (receivedfor review May 1, 1992) ABSTRACT Ethanol is the major metabolic product of NAD(P)+ so that the reducing equivalents are regenerated glucose fermentation by the protozoan parasite Entmoeba (4-7). Both NADP+-dependent and NAD+-dependent alco- histolytica under the anaerobic conditions found in the lumen hol dehydrogenase (ADH) activities have been identified in of the colon. Here an internal peptide sequence determined E. histolytica trophozoites (4-7). The NADP+-dependent from a major 39-kDa amoeba protein isolated by isoeLtric ADH (EhADHi; EC 1.1.1.2) was found to be composed of focusing followed by SDS/PAGE was used to clone the gene for four -30-kDa subunits, prefer secondary alcohols to primary the E. histolytica NADP+-dependent alcohol dehydrogenase alcohols, and be inhibited by the substrate analogue pyrazole (EhADH1; EC 1.1.1.2). The EhADHi clone had an open (7). The amoeba NAD+-dependent ADH (EC 1.1.1.1) is not reading frame that was 360 amino acids long and encoded a well-characterized (4). -

Altered Expression and Function of Mitochondrial Я-Oxidation Enzymes
0031-3998/01/5001-0083 PEDIATRIC RESEARCH Vol. 50, No. 1, 2001 Copyright © 2001 International Pediatric Research Foundation, Inc. Printed in U.S.A. Altered Expression and Function of Mitochondrial -Oxidation Enzymes in Juvenile Intrauterine-Growth-Retarded Rat Skeletal Muscle ROBERT H. LANE, DAVID E. KELLEY, VLADIMIR H. RITOV, ANNA E. TSIRKA, AND ELISA M. GRUETZMACHER Department of Pediatrics, UCLA School of Medicine, Mattel Children’s Hospital at UCLA, Los Angeles, California 90095, U.S.A. [R.H.L.]; and Departments of Internal Medicine [D.E.K., V.H.R.] and Pediatrics [R.H.L., A.E.T., E.M.G.], University of Pittsburgh School of Medicine, Magee-Womens Research Institute, Pittsburgh, Pennsylvania 15213, U.S.A. ABSTRACT Uteroplacental insufficiency and subsequent intrauterine creased in IUGR skeletal muscle mitochondria, and isocitrate growth retardation (IUGR) affects postnatal metabolism. In ju- dehydrogenase activity was unchanged. Interestingly, skeletal venile rats, IUGR alters skeletal muscle mitochondrial gene muscle triglycerides were significantly increased in IUGR skel- expression and reduces mitochondrial NADϩ/NADH ratios, both etal muscle. We conclude that uteroplacental insufficiency alters of which affect -oxidation flux. We therefore hypothesized that IUGR skeletal muscle mitochondrial lipid metabolism, and we gene expression and function of mitochondrial -oxidation en- speculate that the changes observed in this study play a role in zymes would be altered in juvenile IUGR skeletal muscle. To test the long-term morbidity associated with IUGR. (Pediatr Res 50: this hypothesis, mRNA levels of five key mitochondrial enzymes 83–90, 2001) (carnitine palmitoyltransferase I, trifunctional protein of -oxi- dation, uncoupling protein-3, isocitrate dehydrogenase, and mi- Abbreviations tochondrial malate dehydrogenase) and intramuscular triglycer- CPTI, carnitine palmitoyltransferase I ides were quantified in 21-d-old (preweaning) IUGR and control IUGR, intrauterine growth retardation rat skeletal muscle. -

Isocitrate Dehydrogenase 1 (NADP+) (I5036)
Isocitrate Dehydrogenase 1 (NADP+), human recombinant, expressed in Escherichia coli Catalog Number I5036 Storage Temperature –20 °C CAS RN 9028-48-2 IDH1 and IDH2 have frequent genetic alterations in EC 1.1.1.42 acute myeloid leukemia4 and better understanding of Systematic name: Isocitrate:NADP+ oxidoreductase these mutations may lead to an improvement of (decarboxylating) individual cancer risk assessment.6 In addition other studies have shown loss of IDH1 in bladder cancer Synonyms: IDH1, cytosolic NADP(+)-dependent patients during tumor development suggesting this may isocitrate dehydrogenase, isocitrate:NADP+ be involved in tumor progression and metastasis.7 oxidoreductase (decarboxylating), Isocitric Dehydrogenase, ICD1, PICD, IDPC, ICDC, This product is lyophilized from a solution containing oxalosuccinate decarboxylase Tris-HCl, pH 8.0, with trehalose, ammonium sulfate, and DTT. Product Description Isocitrate dehydrogenase (NADP+) [EC 1.1.1.42] is a Purity: ³90% (SDS-PAGE) Krebs cycle enzyme, which converts isocitrate to a-ketoglutarate. The flow of isocitrate through the Specific activity: ³80 units/mg protein glyoxylate bypass is regulated by phosphorylation of isocitrate dehydrogenase, which competes for a Unit definition: 1 unit corresponds to the amount of 1 common substrate (isocitrate) with isocitrate lyase. enzyme, which converts 1 mmole of DL-isocitrate to The activity of the enzyme is dependent on the a-ketoglutarate per minute at pH 7.4 and 37 °C (NADP formation of a magnesium or manganese-isocitrate as cofactor). The activity is measured by observing the 2 complex. reduction of NADP to NADPH at 340 nm in the 7 presence of 4 mM DL-isocitrate and 2 mM MnSO4. -

Pro-Aging Effects of Xanthine Oxidoreductase Products
antioxidants Review Pro-Aging Effects of Xanthine Oxidoreductase Products , , Maria Giulia Battelli y , Massimo Bortolotti y , Andrea Bolognesi * z and Letizia Polito * z Department of Experimental, Diagnostic and Specialty Medicine-DIMES, Alma Mater Studiorum, University of Bologna, Via San Giacomo 14, 40126 Bologna, Italy; [email protected] (M.G.B.); [email protected] (M.B.) * Correspondence: [email protected] (A.B.); [email protected] (L.P.); Tel.: +39-051-20-9-4707 (A.B.); +39-051-20-9-4729 (L.P.) These authors contributed equally. y Co-last authors. z Received: 22 July 2020; Accepted: 4 September 2020; Published: 8 September 2020 Abstract: The senescence process is the result of a series of factors that start from the genetic constitution interacting with epigenetic modifications induced by endogenous and environmental causes and that lead to a progressive deterioration at the cellular and functional levels. One of the main causes of aging is oxidative stress deriving from the imbalance between the production of reactive oxygen (ROS) and nitrogen (RNS) species and their scavenging through antioxidants. Xanthine oxidoreductase (XOR) activities produce uric acid, as well as reactive oxygen and nitrogen species, which all may be relevant to such equilibrium. This review analyzes XOR activity through in vitro experiments, animal studies and clinical reports, which highlight the pro-aging effects of XOR products. However, XOR activity contributes to a regular level of ROS and RNS, which appears essential for the proper functioning of many physiological pathways. This discourages the use of therapies with XOR inhibitors, unless symptomatic hyperuricemia is present. -

RESEARCH COMMUNICATION HADHA Is a Potential Predictor Of
HADHA is a Potential Predictor of the Response to Platinum-based Chemotherapy RESEARCH COMMUNICATION HADHA is a Potential Predictor of Response to Platinum-based Chemotherapy for Lung Cancer Taihei Kageyama1, Ryo Nagashio1, 2, Shinichiro Ryuge 3, Toshihide Matsumoto1,5, Akira Iyoda4, Yukitoshi Satoh4, Noriyuki Masuda3, Shi-Xu Jiang5, Makoto Saegusa5, Yuichi Sato1, 2* Abstract To identify a cisplatin resistance predictor to reduce or prevent unnecessary side effects, we firstly established four cisplatin-resistant sub-lines and compared their protein profiles with cisplatin-sensitive parent lung cancer cell lines using two-dimensional gel electrophoresis. Between the cisplatin-resistant and -sensitive cells, a total of 359 protein spots were differently expressed (>1.5 fold), and 217 proteins (83.0%) were identified. We focused on a mitochondrial protein, hydroxyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase alpha subunit (HADHA), which was increased in all cisplatin-resistant cells. Furthermore, pre- treated biopsy specimens taken from patients who showed resistance to platinum-based treatment showed a significantly higher positive rate for HADHA in all cases (p=0.00367), including non-small cell lung carcinomas (p=0.002), small-cell lung carcinomas (p=0.038), and adenocarcinomas (p=0.008). These results suggest that the expression of HADHA may be a useful marker to predict resistance to platinum-based chemotherapy in patients with lung cancer. Keywords: Cisplatin - HADHA - lung cancer - two-dimensional gel electrophoresis Asian Pacific J Cancer Prev, 12, 3457-3463 Introduction cisplatin resistance rose due to a decrease of blood flow in the tumor and increased DNA repair (Stewart, 2007), Lung cancer is the leading cause of cancer-related the mechanisms underlying cisplatin resistance have not death in the world, and the five-year overall survival rate yet been clarified, and an effective cisplatin resistance is still below 16% (Jemal et al., 2009). -

Glyoxysomal Malate Dehydrogenase from Watermelon Is Synthesized
Proc. Nati. Acad. Sci. USA Vol. 87, pp. 5773-5777, August 1990 Botany Glyoxysomal malate dehydrogenase from watermelon is synthesized with an amino-terminal transit peptide (isoenzymes/organelle/Citrulus vulgaris/polymerase chain reaction) CHRISTINE GIETL* Institute of Botany, Technical University of Munich, Arcisstrasse 16, D-8000 Munich 2, Federal Republic of Germany; and Department of Physiology, Carlsberg Laboratory, Gamle Carlsberg Vej 10, DK-2500 Copenhagen Valby, Denmark Communicated by Diter von Wettstein, May 11, 1990 (receivedfor review March 5, 1990) ABSTRACT The isolation and sequence of a cDNA clone and peroxisomes are seen between mitochondria and chlo- encoding the complete glyoxysomal malate dehydrogenase roplasts (2). As in plants, mammalian peroxisomes contain [gMDH; (S)-malate:NAD+ oxidoreductase, EC 1.1.1.37] of enzymes involved in the production and degradation ofH202; watermelon cotyledons are presented. Partial cDNA clones in trypanosomes, glycolysis is sequestered into microbodies were synthesized in a three part strategy, taking advantage of called glycosomes (3). All microbodies studied contain en- the polymerase chain reaction technology with oligonucleotides zymes for (3-oxidation and a specific spectrum of other based on directly determined amino acid sequences. Subse- enzymes. It is further characteristic that microbody enzyme quently, the complete done for gMDH was synthesized with a activities are also present in other cell compartments. Gly- sense primer corresponding to the nucleotide sequence of the oxysomes as well as mitochondria contain MDH, citrate N-terminal end of pre-gMDIH and an antisense primer corre- synthase, and enzymes for 83-oxidation (4). The organelle- sponding to the adenylylation site found in the mRNA. -

Is There a Role for Ethanol Assay Using Alcohol Dehydrogenase in a Basic Drugs and Alcohol Screen of Blood Following Routine Autopsies?
Journal of Clinical Pathology and Forensic Medicine Vol. 3(2), pp. 17-19, December 2012 Available online at http://www.academicjournals.org/JCPFM DOI: 10.5897/JCPFM12.003 ISSN 2I41-2405 ©2012 Academic Journals Short Communication Is there a role for ethanol assay using alcohol dehydrogenase in a basic drugs and alcohol screen of blood following routine autopsies? William Tormey 1,2 and Tara Moore 3 1Biomedical Sciences, University of Ulster Cromore Road, Coleraine, BT52 1SA, Northern Ireland. 2Department of Chemical Pathology, Beaumont Hospital, Dublin 9, Ireland. 3Biomedical Sciences, University of Ulster, Cromore Road, Coleraine, BT52 1SA, Northern Ireland. Accepted 22 March, 2012 The external examination system of determining the cause of death as operated in Dundee, Scotland is controversial. The addition of a blood or urine specimen for drugs and alcohol processed by hospital autoanalysers will reduce error in death certification. This is even more convenient to organise with a shorter turn-around time than imaging by computed tomography (CT) or magnetic resonance imaging (MRI). These measures may reduce the autopsy rate and ameliorate the distress of families of the deceased. Key words: Autopsy, alcohol, death. INTRODUCTION For over a decade, there has been controversy over the dehydrogenase and the back reaction nicotinamide appropriateness of the high rate of coroner’s autopsies adenine dinucleotide (NAD) to NADH measured (Pounder, 2000; Rutty et al., 2001). Advocacy of the spectrophotometrically at 340 nm on the Olympus 5400 “view and grant” system of issuing the cause of death as autoanalyser for the analysis of alcohol. The data sheet operated in Dundee, Scotland has been challenged and claims that the assay can accurately quantitate alcohol imaging is suggested as a partial answer to lessen the concentrations within the range of 10 to 600 mg/dl. -

Cholic Acid for Treating Inborn Errors of Primary Bile Acid Synthesis NHS England Unique Reference Number URN1623 / NICE ID004
NATIONAL INSTITUTE FOR HEALTH AND CARE EXCELLENCE Clinical evidence review of cholic acid for treating inborn errors of primary bile acid synthesis NHS England unique reference number URN1623 / NICE ID004 Prepared by: NICE on behalf of NHS England Specialised Commissioning About this clinical evidence review Clinical evidence reviews are a summary of the best available evidence for a single technology within a licensed indication, for commissioning by NHS England. The clinical evidence review supports NHS England in producing clinical policies but is not NICE guidance or advice. Summary This evidence review considers cholic acid (Laboratoires CTRS [Orphacol] and Retrophin Europe Ltd [Kolbam]) for treating inborn errors of primary bile acid NICE clinical evidence review of cholic acid for treating inborn errors of primary bile acid synthesis Page 1 of 72 NHS URN1623 NICE ID004 synthesis caused by the following enzyme deficiencies in people aged 1 month and over: 3-beta-hydroxy-delta5-C27-steroid oxidoreductase (3beta-HSD) delta4-3-oxosteroid-5-beta reductase (5beta-reductase) 2- (or alpha-) methylacyl-CoA racemase (AMACR) sterol 27-hydroxylase (presenting as cerebrotendinous xanthomatosis [CTX]) cholesterol 7alpha-hydroxylase (CYP7A1). Inborn errors of primary bile acid synthesis are rare genetic conditions in which enzyme deficiencies prevent the liver from converting cholesterol in the body to bile acids (such as cholic acid and chenodeoxycholic acid). This results in the liver producing high concentrations of atypical (or ‘unusual’) bile acids and intermediary metabolites (some of which are toxic to the liver) in an attempt to establish a normal bile acid pool. Accumulation of potentially toxic atypical bile acids and metabolites, and reduced flow of bile acids may cause liver injury. -
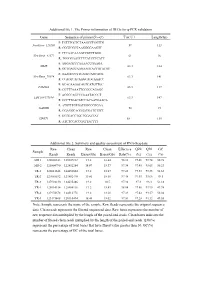
Additional File 1
Additional file 1. The Primer information of DEGs for q-PCR validation Gene Sequence of primer(5'→3') Tm(℃) Length(bp) F: TGTTTGCTCTAAGCCTGGTTG NewGene_126260 59 113 R: CGGTCGCTAAGGGGAAGTT F: CTCAACAAAGCCGTCTGGG NewGene_41572 61 96 R: TGGGGAATCTTCATCCTCATT F: AGGAGCCCAAAACCGAAGA MME 63.3 184 R: GCTGACCAAGAAGTACCGTATGT F: AAAGCCCTTCAGTCAGCACG NewGene_70974 63.3 141 R: CCAGTCACAAGCAGCAAACC F: GCACAAGGCAGTCATGTTGC FAM43A 63.3 117 R: CGTTTAAATTCCGCCAGAGC F: ACGGCAGCCCAAATACCCT LOC108177184 63.3 147 R: GCCTTGACATCCACAATGAACA F: ATGTTTGTGATGGGCGTGAA GAPDH 58 94 R: GGAGGCAGGGATGATGTTCT F: GCTGACCTGCTGGATTAT HPRT1 58 135 R: ATCTCCACCGATTACTTT Additional file 2. Summary and quality assessment of RNA-Seq data Raw Clean Raw Clean Effective Q20 Q30 GC Sample Reads Reads Bases(Gb) Bases(Gb) Rate(%) (%) (%) (%) MR-1 128008342 125607312 19.2 18.84 98.12 97.46 93.74 50.78 MR-2 125804790 122452284 18.87 18.37 97.34 97.45 93.65 50.23 TR-1 128011648 124452604 19.2 18.67 97.22 97.33 93.35 51.65 TR-2 127085532 123882470 19.06 18.58 97.48 97.45 93.66 49.5 TR-3 127994130 124635486 19.2 18.7 97.38 97.3 93.3 51.18 YR-1 128014314 125504116 19.2 18.83 98.04 97.56 93.95 49.98 YR-2 127974678 124512778 19.2 18.68 97.29 97.42 93.57 50.04 YR-3 123173408 120110494 18.48 18.02 97.51 97.28 93.32 49.55 Note: Sample represents the name of the sample. Raw Reads represents the original sequence data. Clean reads represents the filtered sequenced data. Raw bases represents the number of raw sequence data multiplied by the length of the paired-end reads. -

How Is Alcohol Metabolized by the Body?
Overview: How Is Alcohol Metabolized by the Body? Samir Zakhari, Ph.D. Alcohol is eliminated from the body by various metabolic mechanisms. The primary enzymes involved are aldehyde dehydrogenase (ALDH), alcohol dehydrogenase (ADH), cytochrome P450 (CYP2E1), and catalase. Variations in the genes for these enzymes have been found to influence alcohol consumption, alcohol-related tissue damage, and alcohol dependence. The consequences of alcohol metabolism include oxygen deficits (i.e., hypoxia) in the liver; interaction between alcohol metabolism byproducts and other cell components, resulting in the formation of harmful compounds (i.e., adducts); formation of highly reactive oxygen-containing molecules (i.e., reactive oxygen species [ROS]) that can damage other cell components; changes in the ratio of NADH to NAD+ (i.e., the cell’s redox state); tissue damage; fetal damage; impairment of other metabolic processes; cancer; and medication interactions. Several issues related to alcohol metabolism require further research. KEY WORDS: Ethanol-to acetaldehyde metabolism; alcohol dehydrogenase (ADH); aldehyde dehydrogenase (ALDH); acetaldehyde; acetate; cytochrome P450 2E1 (CYP2E1); catalase; reactive oxygen species (ROS); blood alcohol concentration (BAC); liver; stomach; brain; fetal alcohol effects; genetics and heredity; ethnic group; hypoxia The alcohol elimination rate varies state of liver cells. Chronic alcohol con- he effects of alcohol (i.e., ethanol) widely (i.e., three-fold) among individ- sumption and alcohol metabolism are on various tissues depend on its uals and is influenced by factors such as strongly linked to several pathological concentration in the blood T chronic alcohol consumption, diet, age, consequences and tissue damage. (blood alcohol concentration [BAC]) smoking, and time of day (Bennion and Understanding the balance of alcohol’s over time. -

Bioactivity of Curcumin on the Cytochrome P450 Enzymes of the Steroidogenic Pathway
International Journal of Molecular Sciences Article Bioactivity of Curcumin on the Cytochrome P450 Enzymes of the Steroidogenic Pathway Patricia Rodríguez Castaño 1,2, Shaheena Parween 1,2 and Amit V Pandey 1,2,* 1 Pediatric Endocrinology, Diabetology, and Metabolism, University Children’s Hospital Bern, 3010 Bern, Switzerland; [email protected] (P.R.C.); [email protected] (S.P.) 2 Department of Biomedical Research, University of Bern, 3010 Bern, Switzerland * Correspondence: [email protected]; Tel.: +41-31-632-9637 Received: 5 September 2019; Accepted: 16 September 2019; Published: 17 September 2019 Abstract: Turmeric, a popular ingredient in the cuisine of many Asian countries, comes from the roots of the Curcuma longa and is known for its use in Chinese and Ayurvedic medicine. Turmeric is rich in curcuminoids, including curcumin, demethoxycurcumin, and bisdemethoxycurcumin. Curcuminoids have potent wound healing, anti-inflammatory, and anti-carcinogenic activities. While curcuminoids have been studied for many years, not much is known about their effects on steroid metabolism. Since many anti-cancer drugs target enzymes from the steroidogenic pathway, we tested the effect of curcuminoids on cytochrome P450 CYP17A1, CYP21A2, and CYP19A1 enzyme activities. When using 10 µg/mL of curcuminoids, both the 17α-hydroxylase as well as 17,20 lyase activities of CYP17A1 were reduced significantly. On the other hand, only a mild reduction in CYP21A2 activity was observed. Furthermore, CYP19A1 activity was also reduced up to ~20% of control when using 1–100 µg/mL of curcuminoids in a dose-dependent manner. Molecular docking studies confirmed that curcumin could dock onto the active sites of CYP17A1, CYP19A1, as well as CYP21A2. -

1 Here Are the Quiz 4 Questions You Will Answer Online Using the Quiz
Chemistry 6720, quiz 4 handout Here are the quiz 4 questions you will answer online using the quiz tool in canvas instructure. Some of the questions deal with citric acid cycle enzymes that use mechanistic strategies covered in the lectures. Since we did not cover all of those specific enzymes in lecture, be sure to review the citric acid cycle ahead of taking the quiz online so you can answer the appropriate questions regarding mechanistic strategies/themes for the enzymes. In doing so, practice drawing mechanisms for each enzyme of the citric acid cycle. You will probably need to look up the mechanisms for isocitrate dehydrogenase, aconitase, succinate dehydrogenase, and succinyl-CoA synthetase in a general biochemistry text. I will not be asking specific details of the mechanisms for those 4 enzymes, but you will still need to be generally familiar with how they operate to answer a few of the questions below. You do not need to know the chemistry of FAD reduction for the quiz. Be sure to note the quiz deadline in canvas to be sure you get your answers submitted by the deadline. 1. Shown at the right is a Fischer projection for D-glucose. Use the strategy described in the lecture to assign the configurations (R or S) of the chiral centers at C2, C3, C4, and C5. 2. Using the structures shown to the right, assign the methyl groups of isopropanol, and the hydrogens at C2 and C3 of succinate, as either ProR or ProS. Use the strategy covered in lecture to guide you in making the assignments.