Petra Brandt
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hplc-Uv Quantitation of Folate Synthesized by Rickettsia
HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) By Junyan Chen A Thesis Presented to The Faculty of Humboldt State University In Partial Fulfillment of the Requirements for the Degree Master of Science in Biology Committee Membership Dr. Jianmin Zhong, Committee Chair Dr. David S. Baston, Committee Member Dr. Jenny Cappuccio, Committee Member Dr. Jacob Varkey, Committee Member Dr. Erik Jules, Program Graduate Coordinator December 2017 ABSTRACT HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) Junyan Chen Ticks are the most important vector of many infectious diseases in the United States. Understanding the nature of the relationship between Rickettsia endosymbiont Ixodes pacificus (REIP) and Exudes pacificus will help develop strategies for the control of tick- borne diseases, such as Lyme disease, and Rocky Mountain spotted fever. Folate, also known as vitamin B9, is a necessary vitamin for tick survival, and plays a central role in one-carbon metabolism in cells. Folate exist as a large family of structurally related forms that transfer one-carbon groups among biomolecules that are important to cell growth, differentiation, and survival. In Dr. Zheng’s lab, REIP were cultured in Ixodes scapularis embryonic tick cell line ISE6. Previous research has shown that REIP in Ixodes pacificus carries all five de novo folate biosynthesis genes. Folate biosynthesis mRNAs were detected and all recombinant rickettsial folate proteins were overexpressed. To determine whether REIP synthesize folate, we sought to measure the folate concentration in REIP using HPLC-UV quantification with a Diamond HydrideTM liquid chromatography column. 5-methyltetrahydrofolate (5-MTHF), the active circulating form of folate in bacteria was detected. -

Humpback Whale Populations Share a Core Skin Bacterial Community: Towards a Health Index for Marine Mammals?
Humpback Whale Populations Share a Core Skin Bacterial Community: Towards a Health Index for Marine Mammals? Amy Apprill1*, Jooke Robbins2, A. Murat Eren3, Adam A. Pack4,5, Julie Reveillaud3, David Mattila6, Michael Moore1, Misty Niemeyer7, Kathleen M. T. Moore7, Tracy J. Mincer1* 1 Woods Hole Oceanographic Institution, Woods Hole, Massachusetts, United States of America, 2 Center for Coastal Studies, Provincetown, Massachusetts, United States of America, 3 Josephine Bay Paul Center, Marine Biological Laboratory, Woods Hole, Massachusetts, United States of America, 4 University of Hawaii at Hilo, Hilo, Hawaii, United States of America, 5 The Dolphin Institute, Honolulu, Hawaii, United States of America, 6 Hawaiian Islands Humpback Whale National Marine Sanctuary, Kihei, Hawaii, United States of America, 7 International Fund for Animal Welfare, Yarmouth Port, Massachusetts, United States of America Abstract Microbes are now well regarded for their important role in mammalian health. The microbiology of skin – a unique interface between the host and environment - is a major research focus in human health and skin disorders, but is less explored in other mammals. Here, we report on a cross-population study of the skin-associated bacterial community of humpback whales (Megaptera novaeangliae), and examine the potential for a core bacterial community and its variability with host (endogenous) or geographic/environmental (exogenous) specific factors. Skin biopsies or freshly sloughed skin from 56 individuals were sampled from populations in the North Atlantic, North Pacific and South Pacific oceans and bacteria were characterized using 454 pyrosequencing of SSU rRNA genes. Phylogenetic and statistical analyses revealed the ubiquity and abundance of bacteria belonging to the Flavobacteria genus Tenacibaculum and the Gammaproteobacteria genus Psychrobacter across the whale populations. -
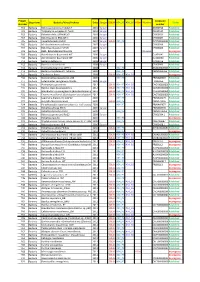
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Diversity of Spotted Fever Group Rickettsiae and Their Association
www.nature.com/scientificreports OPEN Diversity of spotted fever group rickettsiae and their association with host ticks in Japan Received: 31 July 2018 May June Thu1,2, Yongjin Qiu3, Keita Matsuno 4,5, Masahiro Kajihara6, Akina Mori-Kajihara6, Accepted: 14 December 2018 Ryosuke Omori7,8, Naota Monma9, Kazuki Chiba10, Junji Seto11, Mutsuyo Gokuden12, Published: xx xx xxxx Masako Andoh13, Hideo Oosako14, Ken Katakura2, Ayato Takada5,6, Chihiro Sugimoto5,15, Norikazu Isoda1,5 & Ryo Nakao2 Spotted fever group (SFG) rickettsiae are obligate intracellular Gram-negative bacteria mainly associated with ticks. In Japan, several hundred cases of Japanese spotted fever, caused by Rickettsia japonica, are reported annually. Other Rickettsia species are also known to exist in ixodid ticks; however, their phylogenetic position and pathogenic potential are poorly understood. We conducted a nationwide cross-sectional survey on questing ticks to understand the overall diversity of SFG rickettsiae in Japan. Out of 2,189 individuals (19 tick species in 4 genera), 373 (17.0%) samples were positive for Rickettsia spp. as ascertained by real-time PCR amplifcation of the citrate synthase gene (gltA). Conventional PCR and sequencing analyses of gltA indicated the presence of 15 diferent genotypes of SFG rickettsiae. Based on the analysis of fve additional genes, we characterised fve Rickettsia species; R. asiatica, R. helvetica, R. monacensis (formerly reported as Rickettsia sp. In56 in Japan), R. tamurae, and Candidatus R. tarasevichiae and several unclassifed SFG rickettsiae. We also found a strong association between rickettsial genotypes and their host tick species, while there was little association between rickettsial genotypes and their geographical origins. -
Widespread Use of Realtime PCR for Rickettsial Diagnosis
SHORT COMMUNICATION Widespread use of real-time PCR for rickettsial diagnosis Aure´ lie Renvoise´ , Jean-Marc Rolain, Cristina Socolovschi & Didier Raoult Unite´ de Recherche en Maladies Infectieuses et Tropicales Emergentes CNRS-IRD UMR6236-198, Faculte´ de Me´ decine, Universite´ de la Me´ diterrane´ e, Marseille, France Correspondence: Didier Raoult, Unite´ de Abstract Recherche en Maladies Infectieuses et Tropicales Emergentes CNRS-IRD UMR6236- We report 2 years of experience with rickettsial molecular diagnosis using real- 198, Universite´ de la Me´ diterrane´ e, Faculte´ time PCR at the French National Reference Center. All Rickettsia genomes avail- de Me´ decine, 27 bd Jean Moulin, 13385 able were compared to discover specific sequences to design new sets of primers Marseille Cedex 5, France. Tel.: and probes. The specificity was verified in silico and against a panel of 30 rick- +33491324375; fax: +33491387772; ettsial species. Sensitivity was determined using 10-fold serial dilutions. Finally, e-mail: [email protected] primers and probes that were both specific and sensitive were routinely used Received 6 July 2011; revised 6 October for the diagnosis of rickettsial infections from clinical specimens. We retained 2011; accepted 31 October 2011. sets of primers and probes to detect spotted fever group Rickettsia, typhus Final version published online 8 December group Rickettsia, Rickettsia conorii, Rickettsia slovaca, Rickettsia africae and Rick- 2011. ettsia australis; 643 clinical samples were screened for the presence of Rickettsia DNA. Overall, 45 positive samples were detected, including 15 Rickettsia africae, DOI: 10.1111/j.1574-695X.2011.00899.x nine R. conorii, five Rickettsia sibirica mongolitimonae, four R. -

Fish Bacterial Flora Identification Via Rapid Cellular Fatty Acid Analysis
Fish bacterial flora identification via rapid cellular fatty acid analysis Item Type Thesis Authors Morey, Amit Download date 09/10/2021 08:41:29 Link to Item http://hdl.handle.net/11122/4939 FISH BACTERIAL FLORA IDENTIFICATION VIA RAPID CELLULAR FATTY ACID ANALYSIS By Amit Morey /V RECOMMENDED: $ Advisory Committe/ Chair < r Head, Interdisciplinary iProgram in Seafood Science and Nutrition /-■ x ? APPROVED: Dean, SchooLof Fisheries and Ocfcan Sciences de3n of the Graduate School Date FISH BACTERIAL FLORA IDENTIFICATION VIA RAPID CELLULAR FATTY ACID ANALYSIS A THESIS Presented to the Faculty of the University of Alaska Fairbanks in Partial Fulfillment of the Requirements for the Degree of MASTER OF SCIENCE By Amit Morey, M.F.Sc. Fairbanks, Alaska h r A Q t ■ ^% 0 /v AlA s ((0 August 2007 ^>c0^b Abstract Seafood quality can be assessed by determining the bacterial load and flora composition, although classical taxonomic methods are time-consuming and subjective to interpretation bias. A two-prong approach was used to assess a commercially available microbial identification system: confirmation of known cultures and fish spoilage experiments to isolate unknowns for identification. Bacterial isolates from the Fishery Industrial Technology Center Culture Collection (FITCCC) and the American Type Culture Collection (ATCC) were used to test the identification ability of the Sherlock Microbial Identification System (MIS). Twelve ATCC and 21 FITCCC strains were identified to species with the exception of Pseudomonas fluorescens and P. putida which could not be distinguished by cellular fatty acid analysis. The bacterial flora changes that occurred in iced Alaska pink salmon ( Oncorhynchus gorbuscha) were determined by the rapid method. -

Bacterial Diversity in Amblyomma Americanum (Acari: Ixodidae) with Afocusonmembersofthegenusrickettsia
VECTOR-BORNE DISEASES,SURVEILLANCE,PREVENTION Bacterial Diversity in Amblyomma americanum (Acari: Ixodidae) With aFocusonMembersoftheGenusRickettsia 1 2 1 STEPHANIE R. HEISE, M. S. ELSHAHED, AND S. E. LITTLE Department of Veterinary Pathobiology, Center for Veterinary Health Sciences, Oklahoma State University, Stillwater, OK 74078 J. Med. Entomol. 47(2): 258Ð268 (2010); DOI: 10.1603/ME09197 ABSTRACT The lone star tick, Amblyomma americanum (Acari: Ixodidae), is commonly reported from people and animals throughout the eastern U.S. and is associated with transmission of a number of emerging diseases. To better deÞne the microbial communities within lone star ticks, 16S rRNA gene based analysis using bacteria-wide primers, followed by sequencing of individual clones (n ϭ 449) was used to identify the most common bacterial operational taxonomic units (OTUs) present within colony-reared and wild A. americanum.Thecolony-rearedtickscontainedprimarilysequenceafÞl- iated with members of the genus Coxiella (89%; 81/91), common endosymbionts of ticks, and Brevibacterium (11%; 10/91). Similarly, analysis of clones from unfed wild lone star ticks revealed that 96.7% (89/92) of all the OTUs identiÞed were afÞliated with Coxiella-like endosymbionts, as compared with only 5.1Ð11.7% (5/98Ð9/77) of those identiÞed from wild lone star ticks after feeding. In contrast, the proportion of OTUs identiÞed as Rickettsia sp. in wild-caught ticks increased from 2.2% (2/92) before feeding to as high as 46.8% (36/77) after feeding, and all Rickettsia spp. sequences recovered were most similar to those described from the spotted fever group Rickettsia,speciÞcallyR. amblyo- mmii and R. massiliae.AdditionalcharacterizationoftheRickettsialestickcommunitybypolymerase chain reaction, cloning, and sequencing of 17 kDa and gltA genes conÞrmed these initial Þndings and suggested that novel Rickettsia spp. -

Copyright by Casey L. C. Schroeder 2016
Copyright by Casey L. C. Schroeder 2016 Rickettsia prowazekii and its ‘Junk DNA’: The Identification and Characterization of Small RNAs by Casey Lee Cody Schroeder, MS, MLS(ASCP), SM, SLS Dissertation Presented to the Faculty of the Graduate School of The University of Texas Medical Branch in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy The University of Texas Medical Branch October, 2016 Dedication I dedicate this dissertation to my daughter, who has the fortunate pleasure of possessing half of her mother’s genes. Sorry about the other half. Explore. Dream. Discover. For I learned far more in the woods than I did inside a building. Nature is calling. Answer it. Acknowledgments I would like to acknowledge those that were instrumental: -To the Flying Spaghetti Monster: For your noodly appendage has touched and guided my results. Without the logical conjecture based on overwhelming observable evidence, the conclusions in this dissertation would not have been possible. -The Universe: For had you not expanded 13.8 billion years ago and created the Earth 4.5 billion years ago, none of this research would have been physically possible. The continued spewing of chemically-rich star guts atomically binds this research to the universe and biologically to the Earth. For that, I am eternally grateful. -Dissertation Committee: My sympathies for forcing you to sit through, what probably felt like never-ending, biannual dissertation committee meetings with me. If it is any solace, they have ended………. hopefully. Specifically, Dr. Fofanov, you’re fired. Dr. Minnick, enjoy fly fishing with the grizzly bears. -

Ru 2015 150 263 a (51) Мпк A61k 31/155 (2006.01)
РОССИЙСКАЯ ФЕДЕРАЦИЯ (19) (11) (13) RU 2015 150 263 A (51) МПК A61K 31/155 (2006.01) ФЕДЕРАЛЬНАЯ СЛУЖБА ПО ИНТЕЛЛЕКТУАЛЬНОЙ СОБСТВЕННОСТИ (12) ЗАЯВКА НА ИЗОБРЕТЕНИЕ (21)(22) Заявка: 2015150263, 01.05.2014 (71) Заявитель(и): НЕОКУЛИ ПТИ ЛТД (AU) Приоритет(ы): (30) Конвенционный приоритет: (72) Автор(ы): 01.05.2013 AU 2013901517 ПЕЙДЖ Стефен (AU), ГАРГ Санджай (AU) (43) Дата публикации заявки: 06.06.2017 Бюл. № 16 RU (85) Дата начала рассмотрения заявки PCT на национальной фазе: 01.12.2015 (86) Заявка PCT: AU 2014/000480 (01.05.2014) 2015150263 (87) Публикация заявки PCT: WO 2014/176634 (06.11.2014) Адрес для переписки: 190000, Санкт-Петербург, Box-1125, "ПАТЕНТИКА" A (54) СПОСОБЫ ЛЕЧЕНИЯ БАКТЕРИАЛЬНЫХ ИНФЕКЦИЙ (57) Формула изобретения 1. Способ лечения или профилактики бактериальной колонизации или инфекции у субъекта, включающий стадию: введения субъекту терапевтически эффективного количества робенидина или его терапевтически приемлемой соли, причем указанная A бактериальная колонизация или инфекция вызвана бактериальным агентом. 2. Способ по п. 1, отличающийся тем, что субъект выбран из группы, включающей: человека, животных, принадлежащих видам семейства псовых, кошачьих, крупного рогатого скота, овец, коз, свиней, птиц, рыб и лошадей. 3. Способ по п. 1, отличающийся тем, что робенидин вводят субъекту в дозе в диапазоне от 0,1 до 250 мг/кг массы тела. 4. Способ по любому из пп. 1-3, отличающийся тем, что бактериальный агент является 2015150263 грамположительным. 5. Способ по п. 4, отличающийся тем, что бактериальный агент выбран из -

The Microbiome of North Sea Copepods
Helgol Mar Res (2013) 67:757–773 DOI 10.1007/s10152-013-0361-4 ORIGINAL ARTICLE The microbiome of North Sea copepods G. Gerdts • P. Brandt • K. Kreisel • M. Boersma • K. L. Schoo • A. Wichels Received: 5 March 2013 / Accepted: 29 May 2013 / Published online: 29 June 2013 Ó Springer-Verlag Berlin Heidelberg and AWI 2013 Abstract Copepods can be associated with different kinds Keywords Bacterial community Á Copepod Á and different numbers of bacteria. This was already shown in Helgoland roads Á North Sea the past with culture-dependent microbial methods or microscopy and more recently by using molecular tools. In our present study, we investigated the bacterial community Introduction of four frequently occurring copepod species, Acartia sp., Temora longicornis, Centropages sp. and Calanus helgo- Marine copepods may constitute up to 80 % of the meso- landicus from Helgoland Roads (North Sea) over a period of zooplankton biomass (Verity and Smetacek 1996). They are 2 years using DGGE (denaturing gradient gel electrophore- key components of the food web as grazers of primary pro- sis) and subsequent sequencing of 16S-rDNA fragments. To duction and as food for higher trophic levels, such as fish complement the PCR-DGGE analyses, clone libraries of (Cushing 1989; Møller and Nielsen 2001). Copepods con- copepod samples from June 2007 to 208 were generated. tribute to the microbial loop (Azam et al. 1983) due to Based on the DGGE banding patterns of the two years sur- ‘‘sloppy feeding’’ (Møller and Nielsen 2001) and the release vey, we found no significant differences between the com- of nutrients and DOM from faecal pellets (Hasegawa et al. -

Rebecca Olivia Maclennan Cowie a Thesis Submitted to Victoria
BACTERIAL COMMUNITY STRUCTURE, FUNCTION AND DIVERSITY IN ANTARCTIC SEA ICE Rebecca Olivia MacLennan Cowie A thesis submitted to Victoria University of Wellington in fulfillment of the requirement for the degree of Doctor of Philosophy in Ecology & Biodiversity Victoria University of Wellington Te Whare Wananga o te Upoko o te Ika a Maui 2011 “I make no apologies for putting microorganisms on a pedestal above all other living things. For if the last blue whale choked to death on the last panda, it would be disastrous but not the end of the world. But if we accidentally poisoned the last two species of ammonia-oxidisers, that would be another matter. It could be happening now and we wouldn’t even know...” Tom Curtis (July 2006) ACKNOWLEDGEMENTS I would first like to thank my supervisors, Ken Ryan and Els Maas, for without them this thesis would not have been possible. Ken, thank you for everything! Thanks for giving me the opportunity to carry out research as part of the K043 team in Antarctica. I am grateful for the wealth of time you had for me whenever I came knocking on your door. I appreciate for your support, time and effort throughout the last three years and especially during the write-up. Els, thank you for giving so much of your time and energy towards my research. Your knowledge and advice has been invaluable. To my friends, fellow students and office mates who helped me along the way both scientifically and recreationally I thank Bionda Morelissen, Alejandra Perea Blazquez, Charles Lee, David Weller, Abi Powell, Mareike Sudek, Ingrid Knapp and Jade Berman. -

Not Surviving During Air Transport: Evaluation of Microbial Spoilage
Italian Journal of Food Safety 2016; volume 5:5620 American lobsters economic resource of the coastal communities of North America, with more than 130,000 tons Correspondence: Cristian Bernardi, Department (Homarus americanus) of alive animals commercialised worldwide in of Health, Animal Science and Food Safety, not surviving during air 2006 and producing nearly 545 million of euros University of Milan, via G. Celoria 10, 20133 transport: evaluation of profits. Due to the decrease of the European Milan, Italy. lobster Homarus gammarus catching figures, Tel: +39.02.50317855 - Fax: +39.02.50317870. E-mail: [email protected] of microbial spoilage American lobsters are imported by several European countries, especially Mediterranean Erica Tirloni, Simone Stella, Acknowledgements: the authors would like to ones: in Italy about 4387 tons are annually thank METRO Italia SpA for providing the Mario Gennari, Fabio Colombo, introduced for an economic value of 44 million American lobsters. Professor Patrizia Cattaneo Cristian Bernardi of euros (Barrento et al., 2009; FAO, 2006). should also be acknowledged for her careful revi- Department of Health, Animal Science American lobsters are mainly commercialised sion of the manuscript. alive and cooked before consumption. After the and Food Safety, University of Milan, Key words: Dead American lobsters; Shipping; capture and during the air transport, they are Milan, Italy Food spoilage; Microbial population. submitted to several stresses like air exposure, changes of the natural environments in terms Contributions: ET and SS were involved in micro- of physical and chemical parameters such as biological analyses and writing of the paper. FC Abstract water salinity and temperature, hypoxia, fast- was involved in biomolecular analyses.