The Unfolded Protein Response and Its Potential Role In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

CCT2 Antibody A
Revision 5 C 0 2 - t CCT2 Antibody a e r o t S Orders: 877-616-CELL (2355) [email protected] Support: 877-678-TECH (8324) 1 6 Web: [email protected] 5 www.cellsignal.com 3 # 3 Trask Lane Danvers Massachusetts 01923 USA For Research Use Only. Not For Use In Diagnostic Procedures. Applications: Reactivity: Sensitivity: MW (kDa): Source: UniProt ID: Entrez-Gene Id: WB, IP H M R Mk Endogenous 54 Rabbit P78371 10576 Product Usage Information 6. McCormack, E.A. et al. (2001) J Struct Biol 135, 198-204. Application Dilution Western Blotting 1:1000 Immunoprecipitation 1:100 Storage Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA and 50% glycerol. Store at –20°C. Do not aliquot the antibody. Specificity / Sensitivity CCT2 Antibody detects endogenous levels of total CCT2 protein. Species Reactivity: Human, Mouse, Rat, Monkey Source / Purification Polyclonal antibodies are produced by immunizing animals with a synthetic peptide corresponding to human CCT2. Antibodies are purified by protein A and peptide affinity chromatography. Background CCT2 is one of eight largely unrelated subunit proteins found in a protein chaperone complex known as the chaperonin-containing TCP-1 (CCT) or TRiC complex. The CCT complex is an abundanct cytoslic component that is credited with helping newly synthesized polypeptides adopt the correct conformation (1). Proteins that fold and assemble with the help of CCT include the cytoskeletal proteins actin and tubulin as well as up to 15% of newly synthesized eukaryotic proteins (2). CCT2 is the β-subunit of the chaperone complex and is one of several CCT proteins that exhibit increased expression in response to stress. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -

Proteomic and Bioinformatic Investigation of Altered Pathways in Neuroglobin-Deficient Breast Cancer Cells
molecules Article Proteomic and Bioinformatic Investigation of Altered Pathways in Neuroglobin-Deficient Breast Cancer Cells Michele Costanzo 1,2 , Marco Fiocchetti 3 , Paolo Ascenzi 3, Maria Marino 3 , Marianna Caterino 1,2,* and Margherita Ruoppolo 1,2,* 1 Department of Molecular Medicine and Medical Biotechnology, School of Medicine, University of Naples Federico II, 80131 Naples, Italy; [email protected] 2 CEINGE—Biotecnologie Avanzate S.C.Ar.L., 80145 Naples, Italy 3 Department of Science, University Roma Tre, 00146 Rome, Italy; marco.fi[email protected] (M.F.); [email protected] (P.A.); [email protected] (M.M.) * Correspondence: [email protected] (M.C.); [email protected] (M.R.) Abstract: Neuroglobin (NGB) is a myoglobin-like monomeric globin that is involved in several processes, displaying a pivotal redox-dependent protective role in neuronal and extra-neuronal cells. NGB remarkably exerts its function upon upregulation by NGB inducers, such as 17β-estradiol (E2) and H2O2. However, the molecular bases of NGB’s functions remain undefined, mainly in non- neuronal cancer cells. Human MCF-7 breast cancer cells with a knocked-out (KO) NGB gene obtained using CRISPR/Cas9 technology were analyzed using shotgun label-free quantitative proteomics in comparison with control cells. The differential proteomics experiments were also performed after treatment with E2, H2O2, and E2 + H2O2. All the runs acquired using liquid chromatography–tandem mass spectrometry were elaborated within the same MaxQuant analysis, leading to the quantification Citation: Costanzo, M.; Fiocchetti, of 1872 proteins in the global proteomic dataset. Then, a differentially regulated protein dataset was M.; Ascenzi, P.; Marino, M.; Caterino, M.; Ruoppolo, M. -

Supplementary Table S1. Correlation Between the Mutant P53-Interacting Partners and PTTG3P, PTTG1 and PTTG2, Based on Data from Starbase V3.0 Database
Supplementary Table S1. Correlation between the mutant p53-interacting partners and PTTG3P, PTTG1 and PTTG2, based on data from StarBase v3.0 database. PTTG3P PTTG1 PTTG2 Gene ID Coefficient-R p-value Coefficient-R p-value Coefficient-R p-value NF-YA ENSG00000001167 −0.077 8.59e-2 −0.210 2.09e-6 −0.122 6.23e-3 NF-YB ENSG00000120837 0.176 7.12e-5 0.227 2.82e-7 0.094 3.59e-2 NF-YC ENSG00000066136 0.124 5.45e-3 0.124 5.40e-3 0.051 2.51e-1 Sp1 ENSG00000185591 −0.014 7.50e-1 −0.201 5.82e-6 −0.072 1.07e-1 Ets-1 ENSG00000134954 −0.096 3.14e-2 −0.257 4.83e-9 0.034 4.46e-1 VDR ENSG00000111424 −0.091 4.10e-2 −0.216 1.03e-6 0.014 7.48e-1 SREBP-2 ENSG00000198911 −0.064 1.53e-1 −0.147 9.27e-4 −0.073 1.01e-1 TopBP1 ENSG00000163781 0.067 1.36e-1 0.051 2.57e-1 −0.020 6.57e-1 Pin1 ENSG00000127445 0.250 1.40e-8 0.571 9.56e-45 0.187 2.52e-5 MRE11 ENSG00000020922 0.063 1.56e-1 −0.007 8.81e-1 −0.024 5.93e-1 PML ENSG00000140464 0.072 1.05e-1 0.217 9.36e-7 0.166 1.85e-4 p63 ENSG00000073282 −0.120 7.04e-3 −0.283 1.08e-10 −0.198 7.71e-6 p73 ENSG00000078900 0.104 2.03e-2 0.258 4.67e-9 0.097 3.02e-2 Supplementary Table S2. -

Genome-Wide Methylation Study on Depression: Differential Methylation and Variable Methylation in Monozygotic Twins
OPEN Citation: Transl Psychiatry (2015) 5, e557; doi:10.1038/tp.2015.49 www.nature.com/tp ORIGINAL ARTICLE Genome-wide methylation study on depression: differential methylation and variable methylation in monozygotic twins A Córdova-Palomera1,2, M Fatjó-Vilas1,2, C Gastó2,3,4, V Navarro2,3,4, M-O Krebs5,6,7 and L Fañanás1,2 Depressive disorders have been shown to be highly influenced by environmental pathogenic factors, some of which are believed to exert stress on human brain functioning via epigenetic modifications. Previous genome-wide methylomic studies on depression have suggested that, along with differential DNA methylation, affected co-twins of monozygotic (MZ) pairs have increased DNA methylation variability, probably in line with theories of epigenetic stochasticity. Nevertheless, the potential biological roots of this variability remain largely unexplored. The current study aimed to evaluate whether DNA methylation differences within MZ twin pairs were related to differences in their psychopathological status. Data from the Illumina Infinium HumanMethylation450 Beadchip was used to evaluate peripheral blood DNA methylation of 34 twins (17 MZ pairs). Two analytical strategies were used to identify (a) differentially methylated probes (DMPs) and (b) variably methylated probes (VMPs). Most DMPs were located in genes previously related to neuropsychiatric phenotypes. Remarkably, one of these DMPs (cg01122889) was located in the WDR26 gene, the DNA sequence of which has been implicated in major depressive disorder from genome-wide association studies. Expression of WDR26 has also been proposed as a biomarker of depression in human blood. Complementarily, VMPs were located in genes such as CACNA1C, IGF2 and the p38 MAP kinase MAPK11, showing enrichment for biological processes such as glucocorticoid signaling. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Kif1b Rab7a Lmna
Title: Charcot-Marie-Tooth Neuropathy Type 2 GeneReview Molecular Genetics: Less Commonly Involved Genes Author: Bird TD Updated: March 2016 KIF1B Gene structure. KIF1B comprises 47 exons and 167.13 kb of DNA. Pathogenic allelic variants. See Table A, Locus Specific and HGMD Normal gene product. Kinesin-like protein KIF1B is involved in axonal transport of synaptic vesicle precursors [Zhao et al 2001]. The kinesin superfamily of proteins is essential for intracellular transport along microtubules. Abnormal gene product. There may be a defect in the transport of synaptic vesicles. RAB7A Gene structure. RAB7A has six exons and 87.9 kb of DNA. Pathogenic allelic variants. See Table A. Normal gene product. Ras-related protein Rab-7a belongs to the RAB family of Ras- related GTPases essential for the regulation of intracellular membrane trafficking. Rab- 7a is involved in transport between late endosomes and lysosomes. RAB-interacting lysosomal protein (RILP) induces the recruitment of dynein-dynactin motors and regulates transport toward the minus-end of microtubules [Verhoeven et al 2003]. Abnormal gene product. Abnormal Rab-7a may cause malfunction of lysosomes and inhibit neurite outgrowth [Spinosa et al 2008, Bucci & Deluca 2012]. LMNA Gene structure. LMNA has 12 exons spread over 24 kb of genomic DNA. Pathogenic allelic variants. The most common pathogenic variant found in individuals with CMT2B1 is p.Arg298Cys, a founder mutation in North Africa [Bouhouche et al 2007, De Sandre-Giovannoli et al 2002]. See also Table A. Table 5. Selected LMNA Variants DNA Nucleotide Protein Amino Acid Class of Variant Allele Reference Sequences Change Change Benign c.1908C>T p.= 1 c.398G>T p.Arg133Leu NM_170707.2 c.892C>T p.Arg298Cys Pathogenic NP_733821.1 c.1411C>T p.Arg471Cys c.1579C>T p.Arg527Cys Note on variant classification: Variants listed in the table have been provided by the author. -

Maser: One-Stop Platform for NGS Big Data from Analysis to Visualization
Database, 2018, 1–12 doi: 10.1093/database/bay027 Database tool Database tool Maser: one-stop platform for NGS big data from analysis to visualization Sonoko Kinjo1, Norikazu Monma2, Sadahiko Misu3, Norikazu Kitamura1, Junichi Imoto1, Kazutoshi Yoshitake4, Takashi Gojobori5 and Kazuho Ikeo1,6,* 1Center for Information Biology, National Institute of Genetics, 1111 Yata, Mishima, Shizuoka 411-8540, Japan, 2Hitachi, Ltd, Tokyo, Japan, 3BITS Co., Ltd, Tokyo, Japan, 4Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan, 5Computational Bioscience Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia and 6Department of Genetics, SOKENDAI, Mishima, Japan *Corresponding author: Tel: þ81 55 981 6851; Fax: þ81 55 981 6848; E-mail: [email protected] Citation details: Kinjo,S., Monma,N., Misu,S. et al. Maser: one-stop platform for NGS big data from analysis to visualiza- tion. Database (2018) Vol. 2018: article ID bay027; doi:10.1093/database/bay027 Received 15 September 2017; Revised 22 February 2018; Accepted 26 February 2018 Abstract A major challenge in analyzing the data from high-throughput next-generation sequenc- ing (NGS) is how to handle the huge amounts of data and variety of NGS tools and visu- alize the resultant outputs. To address these issues, we developed a cloud-based data analysis platform, Maser (Management and Analysis System for Enormous Reads), and an original genome browser, Genome Explorer (GE). Maser enables users to manage up to 2 terabytes of data to conduct analyses with easy graphical user interface operations and offers analysis pipelines in which several individual tools are combined as a single pipeline for very common and standard analyses. -
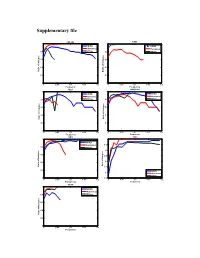
Supplementary File
Supplementary file COL1A2 FZD3 1 1 HPRD HPRD Humannet Humannet 0.8 PPIwu 0.8 PPIwu 0.6 0.6 0.4 0.4 Ratio of of indegree Ratio Ratio of Indegree 0.2 0.2 0 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency RBL1 SMARCA4 1 1 HPRD HPRD Humannet Humannet 0.8 PPIwu 0.8 PPIwu 0.6 0.6 0.4 0.4 Ratio of Indegree Ratio of Indegree Ratio 0.2 0.2 0 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency SRC TP53 1 1 HPRD Humannet 0.95 0.8 PPIwu 0.9 e 0.6 0.85 0.8 0.4 Ratio of Indegree of Ratio Ratio of Indegre 0.75 0.2 HPRD 0.7 Humannet PPIwu 0 0.65 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency VCAN 1 HPRD Humannet 0.8 PPIwu 0.6 0.4 Ratio of Indegree 0.2 0 0 0.05 0.1 0.15 0.2 Frequency Figure S1. Variation of indegree ratio with frequency cutoffs from 0 to 0.2 with a step 0.002 for driver genes common by three fitness cores in COAD. The indegree ratio is set to NULL if total degree of this genes is less than 5. BRAF CASR 1 1 0.8 0.8 0.6 0.6 0.4 0.4 Ratio of Indegree Ratio of Indegree of Ratio HPRD HPRD 0.2 0.2 Humannet Humannet PPIwu PPIwu 0 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency HDAC9 NRAS 1 1 0.8 0.8 0.6 0.6 0.4 0.4 HPRD HPRD Ratio of Indegree Ratio of Indegree 0.2 0.2 Humannet Humannet PPIwu PPIwu 0 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency NF1 CNTNAP2 1 1 0.9 0.8 0.8 0.6 0.7 0.4 HPRD HPRD Ratio of Indegree Ratio of Indegree 0.6 Humannet 0.2 Humannet PPIwu PPIwu 0.5 0 0 0.05 0.1 0.15 0.2 0 0.05 0.1 0.15 0.2 Frequency Frequency Figure S2. -

A Master Autoantigen-Ome Links Alternative Splicing, Female Predilection, and COVID-19 to Autoimmune Diseases
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.30.454526; this version posted August 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. A Master Autoantigen-ome Links Alternative Splicing, Female Predilection, and COVID-19 to Autoimmune Diseases Julia Y. Wang1*, Michael W. Roehrl1, Victor B. Roehrl1, and Michael H. Roehrl2* 1 Curandis, New York, USA 2 Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, USA * Correspondence: [email protected] or [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.30.454526; this version posted August 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Chronic and debilitating autoimmune sequelae pose a grave concern for the post-COVID-19 pandemic era. Based on our discovery that the glycosaminoglycan dermatan sulfate (DS) displays peculiar affinity to apoptotic cells and autoantigens (autoAgs) and that DS-autoAg complexes cooperatively stimulate autoreactive B1 cell responses, we compiled a database of 751 candidate autoAgs from six human cell types. At least 657 of these have been found to be affected by SARS-CoV-2 infection based on currently available multi-omic COVID data, and at least 400 are confirmed targets of autoantibodies in a wide array of autoimmune diseases and cancer.