Patronus Is the Elusive Plant Securin, Preventing Chromosome Separation By
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
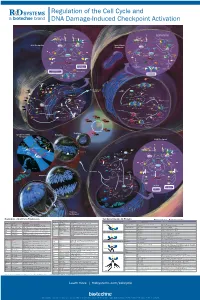
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

Control of the Cell Cycle During Meiotic Maturation of Mouse Oocytes
Control of the cell cycle during meiotic maturation of mouse oocytes Ibtissem Nabti Department of Cell and Developmental Biology University College London London, UK A thesis submitted for the degree of Doctor of Philosophy April 2010 Declaration The work presented in this thesis was conducted under the supervision of Professor John Carroll in the department of cell and developmental biology, University College London, and Professor Keith Jones in the department of reproductive physiology, University of Newcastle upon Tyne. I, Ibtissem Nabti confirm that the data presented in this thesis is my own and from original studies. Also, this dissertation has not previously been submitted, wholly or in part, to any other university for any degree or diploma. Ibtissem Nabti 1 Abstract The overall aim of the work presented in this thesis is to better understand the molecular mechanisms underlying the progression of the meiotic division in mammalian oocytes. The first goal of this project was to investigate the relative contribution of securin and cyclin B1 to the control of the protease separase during the indeterminate period of metaphase II arrest. I found here that although there are conditions in which either securin or MPF can prevent chromosome disjunction, such as during meiosis I, securin is the predominant inhibitor of separase during metaphase II arrest. Thus, CDK1 pharmacological inhibition as well as antibody inhibition of CDK1/cyclin B1 binding to separase, both failed to induce sister chromatid disjunction. Instead, securin morpholino knockdown induced sister chromatid separation, which could be rescued by injection of securin cRNA. I also examined the effect of CDK1 and MAPK activities on securin stability. -

APC/C-Cdh1-Dependent Anaphase and Telophase Progression During
Toda et al. Cell Division 2012, 7:4 http://www.celldiv.com/content/7/1/4 RESEARCH Open Access APC/C-Cdh1-dependent anaphase and telophase progression during mitotic slippage Kazuhiro Toda1, Kayoko Naito1, Satoru Mase1, Masaru Ueno1,2, Masahiro Uritani1, Ayumu Yamamoto1 and Takashi Ushimaru1* Abstract Background: The spindle assembly checkpoint (SAC) inhibits anaphase progression in the presence of insufficient kinetochore-microtubule attachments, but cells can eventually override mitotic arrest by a process known as mitotic slippage or adaptation. This is a problem for cancer chemotherapy using microtubule poisons. Results: Here we describe mitotic slippage in yeast bub2Δ mutant cells that are defective in the repression of precocious telophase onset (mitotic exit). Precocious activation of anaphase promoting complex/cyclosome (APC/ C)-Cdh1 caused mitotic slippage in the presence of nocodazole, while the SAC was still active. APC/C-Cdh1, but not APC/C-Cdc20, triggered anaphase progression (securin degradation, separase-mediated cohesin cleavage, sister- chromatid separation and chromosome missegregation), in addition to telophase onset (mitotic exit), during mitotic slippage. This demonstrates that an inhibitory system not only of APC/C-Cdc20 but also of APC/C-Cdh1 is critical for accurate chromosome segregation in the presence of insufficient kinetochore-microtubule attachments. Conclusions: The sequential activation of APC/C-Cdc20 to APC/C-Cdh1 during mitosis is central to accurate mitosis. Precocious activation of APC/C-Cdh1 in metaphase (pre-anaphase) causes mitotic slippage in SAC-activated cells. For the prevention of mitotic slippage, concomitant inhibition of APC/C-Cdh1 may be effective for tumor therapy with mitotic spindle poisons in humans. -

Functional Interaction Between Bubr1 and Securin in an Anaphase- Promoting Complex/Cyclosomecdc20–Independent Manner
Research Article Functional Interaction between BubR1 and Securin in an Anaphase- Promoting Complex/CyclosomeCdc20–Independent Manner Hyun-Soo Kim,1 Yoon-Kyung Jeon,2 Geun-Hyoung Ha,1 Hye-Young Park,1 Yu-Jin Kim,1 Hyun-Jin Shin,1 Chang Geun Lee,1 Doo-Hyun Chung,2 and Chang-Woo Lee1 1Department of Molecular Cell Biology, Center for Molecular Medicine, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Suwon, Gyeonggi, Korea and 2Department of Pathology, Seoul National University College of Medicine, Seoul, Korea Abstract Cdc20 (APC/CCdc20). However, Cdc20 in MCC is unable to acti- vate APC/C, as shown by several experiments in which the addi- Activation of the mitotic checkpoint requires the precise tion of Mad2 and/or BubR1 leads to the checkpoint-mediated timing and spatial organization of mitotic regulatory inhibition of APC/CCdc20 activity (2, 10, 11). Silencing of this events, and ensures accurate chromosome segregation. checkpoint signal is initiated by the release of the inhibitory Mitotic checkpoint proteins such as BubR1 and Mad2 bind mitotic checkpoint protein complex from APC/CCdc20; APC/CCdc20 to Cdc20, and inhibit anaphase-promoting complex/cyclo- can drive cells into anaphase by inducing the degradation of someCdc20–mediated securin degradation and the onset of securin (pituitary tumor-transforming gene, PTTG), a small protein anaphase. BubR1 mediates the proper attachment of micro- that inhibits the protease separase and mitotic cyclins (12–15). tubules to kinetochores, and links the regulation of chromo- In metaphase-to-anaphase transition, APC/CCdc20 initiates the some-spindle attachment to mitotic checkpoint signaling. process of chromosome segregation through ubiquitination of Therefore, disruption of BubR1 activity results in a loss securin. -

CDK Regulation of Meiosis: Lessons from S. Cerevisiae and S. Pombe
G C A T T A C G G C A T genes Review CDK Regulation of Meiosis: Lessons from S. cerevisiae and S. pombe Anne M. MacKenzie and Soni Lacefield * Department of Biology, Indiana University, 1001 E. Third Street, Bloomington, IN 47405, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-812-856-2429 Received: 16 May 2020; Accepted: 26 June 2020; Published: 29 June 2020 Abstract: Meiotic progression requires precise orchestration, such that one round of DNA replication is followed by two meiotic divisions. The order and timing of meiotic events is controlled through the modulation of the phosphorylation state of proteins. Key components of this phospho-regulatory system include cyclin-dependent kinase (CDK) and its cyclin regulatory subunits. Over the past two decades, studies in budding and fission yeast have greatly informed our understanding of the role of CDK in meiotic regulation. In this review, we provide an overview of how CDK controls meiotic events in both budding and fission yeast. We discuss mechanisms of CDK regulation through post-translational modifications and changes in the levels of cyclins. Finally, we highlight the similarities and differences in CDK regulation between the two yeast species. Since CDK and many meiotic regulators are highly conserved, the findings in budding and fission yeasts have revealed conserved mechanisms of meiotic regulation among eukaryotes. Keywords: meiosis; Cyclin-Dependent Kinase; CDK; cyclin; APC/C; budding yeast; fission yeast; chromosome segregation 1. Introduction Control of the eukaryotic cell cycle occurs through the modulation of phosphorylation states of proteins that trigger specific events. At the forefront of this phospho-regulation are the cyclin-dependent kinases (CDKs), whose oscillatory activity results in a large number of phosphorylations that change the activation state of their substrates [1,2]. -

Structural Analysis of Human Cdc20 Supports Multisite Degron Recognition by APC/C
Structural analysis of human Cdc20 supports multisite degron recognition by APC/C Wei Tiana,1, Bing Lia,b,1, Ross Warringtona,b, Diana R. Tomchickc, Hongtao Yua,b,2, and Xuelian Luoa,2 aDepartment of Pharmacology, bHoward Hughes Medical Institute, and cDepartment of Biophysics, University of Texas Southwestern Medical Center, Dallas, TX 75390 Edited by Stephen C. Harrison, Howard Hughes Medical Institute and Children’s Hospital, Harvard Medical School, Boston, MA, and approved September 24, 2012 (received for review August 3, 2012) The anaphase-promoting complex/cyclosome (APC/C) promotes suggested to inhibit APC/C by acting as a pseudosubstrate, with anaphase onset and mitotic exit through ubiquitinating securin its KEN boxes competing against those of the substrates for and cyclin B1. The mitotic APC/C activator, the cell division cycle 20 Cdc20 binding (22, 23). (Cdc20) protein, directly interacts with APC/C degrons––the de- To better understand how APC/C recognizes its substrates, struction (D) and KEN boxes. APC/CCdc20 is the target of the spindle we determined the crystal structures of human Cdc20 alone or checkpoint. Checkpoint inhibition of APC/CCdc20 requires the bind- bound to the KEN1 box of BubR1, revealing a preexisting KEN- box-binding site at the top of the WD40 β propeller. Structure- ing of a BubR1 KEN box to Cdc20. How APC/C recognizes sub- based mutagenesis then defined a second conserved surface on strates is not understood. We report the crystal structures of the side of the β propeller as a D-box-contacting site. Surpris- human Cdc20 alone or bound to a BubR1 KEN box. -

Inhibitory Factors Associated with Anaphase-Promoting Complex/Cylosome in Mitotic Checkpoint
Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint Ilana Braunstein, Shirly Miniowitz, Yakir Moshe, and Avram Hershko* Unit of Biochemistry, The Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Haifa 31096, Israel Contributed by Avram Hershko, January 19, 2007 (sent for review December 20, 2006) The mitotic (or spindle assembly) checkpoint system ensures accu- checkpoint proteins Mad1, Mad2, and Bub3, protein kinases rate chromosome segregation by preventing anaphase initiation Bub1, BubR1, and Mps1, as well as kinetochore proteins such as until all chromosomes are correctly attached to the mitotic spindle. CENP-E. Most checkpoint proteins and protein kinases are It affects the activity of the anaphase-promoting complex/cyclo- concentrated on unoccupied kinetochores when the checkpoint some (APC/C), a ubiquitin ligase that targets inhibitors of anaphase is active, suggesting that some of the important interactions initiation for degradation. The mechanisms by which this system between these proteins may take place at the kinetochores regulates APC/C remain obscure. Some models propose that the (reviewed in refs. 1–4). system promotes sequestration of the APC/C activator Cdc20 by Although these studies, employing approaches of yeast genet- binding to the checkpoint proteins Mad2 and BubR1. A different ics and mammalian cell biology, were very important in describ- model suggests that a mitotic checkpoint complex (MCC) composed ing the properties and identifying many components of this of BubR1, Bub3, Cdc20, and Mad2 inhibits APC/C in mitotic check- checkpoint system, they could not provide sufficient information point [Sudakin V, Chan GKT, Yen TJ (2001) J Cell Biol 154:925–936]. -

RCS1, a Substrate of APC/C, Controls the Metaphase to Anaphase Transition
RCS1, a substrate of APC/C, controls the metaphase to anaphase transition Wei-meng Zhao*, Judith A. Coppinger†, Akiko Seki*, Xiao-li Cheng*, John R. Yates III†, and Guowei Fang*‡ *Department of Biological Sciences, Stanford University, Stanford, CA 94305-5020; and †Department of Chemical Physiology, The Scripps Research Institute, La Jolla, CA 92037 Edited by Stephen J. Elledge, Harvard Medical School, Boston, MA, and approved June 26, 2008 (received for review September 28, 2007) The anaphase-promoting complex/cyclosome (APC/C) controls the ing of its function in the cell cycle. We report here the identification onset of anaphase by targeting securin for destruction. We report of a substrate of APC/C through a powerful functional genomic here the identification and characterization of a substrate of APC/C, analysis (5) and the characterization of the physiological function of RCS1, as a mitotic regulator that controls the metaphase-to-anaphase this substrate at the metaphase-to-anaphase transition. transition. We showed that the levels of RCS1 fluctuate in the cell cycle, peaking in mitosis and dropping drastically as cells exit into G1. Results Indeed, RCS1 is efficiently ubiquitinated by APC/C in vitro and de- Functional Genomic Analyses Identified Substrates of APC/C. To graded during mitotic exit in a Cdh1-dependent manner in vivo. develop a systematic approach for the identification of APC/C APC/C recognizes a unique D-box at the N terminus of RCS1, as substrates (5), we analyzed 16 known substrates of the APC/C that mutations of this D-box abolished ubiquitination in vitro and stabi- have been characterized at the onset of this study. -

DNA Damage-Induced Mitotic Catastrophe Is Mediated by the Chk1-Dependent Mitotic Exit DNA Damage Checkpoint
DNA damage-induced mitotic catastrophe is mediated by the Chk1-dependent mitotic exit DNA damage checkpoint Xingxu Huang*, Thanh Tran*, Lingna Zhang*, Rashieda Hatcher*, and Pumin Zhang*†‡ *Departments of Molecular Physiology and Biophysics and †Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030 Communicated by Stephen J. Elledge, Harvard Medical School, Boston, MA, December 8, 2004 (received for review November 17, 2004) Mitotic catastrophe is the response of mammalian cells to mitotic so-formed binucleated cells are arrested in G1 in a p53-dependent DNA damage. It produces tetraploid cells with a range of different manner (12, 13). nuclear morphologies from binucleated to multimicronucleated. In It is unclear whether mammalian cells maintain a metaphase response to DNA damage, checkpoints are activated to delay cell DNA damage checkpoint that prevents securin degradation, as in cycle progression and to coordinate repair. Cells in different cell budding yeast, and whether DNA damage-induced mitotic catas- cycle phases use different mechanisms to arrest their cell cycle trophe is regulated by DNA damage checkpoints. We report here progression. It has remained unclear whether the termination of that, in contrast to budding yeast, mammalian securin is not mitosis in a mitotic catastrophe is regulated by DNA damage stabilized in response to mitotic DNA damage. Instead, there is a checkpoints. Here, we report the presence of a mitotic exit DNA mitotic exit DNA damage checkpoint that delays mitotic exit and damage checkpoint in mammalian cells. This checkpoint delays prevents cytokinesis, resulting in the formation of binucleated cells. mitotic exit and prevents cytokinesis and, thereby, is responsible for mitotic catastrophe. -

Regulation of Mitotic Exit by Cell Cycle Checkpoints: Lessons from Saccharomyces Cerevisiae
G C A T T A C G G C A T genes Review Regulation of Mitotic Exit by Cell Cycle Checkpoints: Lessons From Saccharomyces cerevisiae Laura Matellán and Fernando Monje-Casas * Centro Andaluz de Biología Molecular y Medicina Regenerativa (CABIMER), Spanish National Research Council (CSIC)—University of Seville—University Pablo de Olavide, Avda, Américo Vespucio, 24, 41092 Sevilla, Spain; [email protected] * Correspondence: [email protected] Received: 14 January 2020; Accepted: 11 February 2020; Published: 12 February 2020 Abstract: In order to preserve genome integrity and their ploidy, cells must ensure that the duplicated genome has been faithfully replicated and evenly distributed before they complete their division by mitosis. To this end, cells have developed highly elaborated checkpoints that halt mitotic progression when problems in DNA integrity or chromosome segregation arise, providing them with time to fix these issues before advancing further into the cell cycle. Remarkably, exit from mitosis constitutes a key cell cycle transition that is targeted by the main mitotic checkpoints, despite these surveillance mechanisms being activated by specific intracellular signals and acting at different stages of cell division. Focusing primarily on research carried out using Saccharomyces cerevisiae as a model organism, the aim of this review is to provide a general overview of the molecular mechanisms by which the major cell cycle checkpoints control mitotic exit and to highlight the importance of the proper regulation of this process for the maintenance of genome stability during the distribution of the duplicated chromosomes between the dividing cells. Keywords: mitosis; checkpoint; DNA damage; chromosome segregation; aneuploidy 1. -

Structural Analysis of Bub3 Interactions in the Mitotic Spindle Checkpoint
Structural analysis of Bub3 interactions in the mitotic spindle checkpoint Nicholas A. Larsen*, Jawdat Al-Bassam*, Ronnie R. Wei*†, and Stephen C. Harrison*†‡ *Jack Eileen Connors Structural Biology Laboratory, and †Howard Hughes Medical Institute, Harvard Medical School, 250 Longwood Avenue, Boston, MA 02115 Contributed by Stephen C. Harrison, November 27, 2006 (sent for review November 2, 2006) The Mad3/BubR1, Mad2, Bub1, and Bub3 proteins are gatekeepers difference between Mad3 and BubR1 is the retention of the for the transition from metaphase to anaphase. Mad3 from Sac- kinase domain in higher eukaryotes. This kinase domain is charomyces cerevisiae has homology to Bub1 but lacks a corre- activated by the microtubule-associated protein CENP-E, which sponding C-terminal kinase domain. Mad3 forms a stable het- has no homolog in lower eukaryotes (9). erodimer with Bub3. Negative-stain electron microscopy shows Mad3 and the corresponding domain in Bub1 have high helical that Mad3 is an extended molecule (Ϸ200 Å long), whereas Bub3 content and may contain tetracotripeptide repeats (10, 11). Dele- is globular. The Gle2-binding-sequence (GLEBS) motifs found in tion mapping and pull-down experiments have shown that the Mad3 and Bub1 are necessary and sufficient for interaction with Mad3–Bub3 and Bub1–Bub3 interactions are probably restricted to Bub3. The calorimetrically determined dissociation constants for a conserved Gle2-binding sequence (GLEBS) motif (Fig. 1A) (12, GLEBS-motif peptides and Bub3 are Ϸ5 M. Crystal structures of 13). The GLEBS motif was first characterized in the nuclear pore these peptides with Bub3 show that the interactions for Mad3 and complex protein Nup98 and found to be sufficient for binding the Bub1 are similar and mutually exclusive. -

Bimodal Activation of Bubr1 by Bub3 Sustains Mitotic Checkpoint Signaling
Bimodal activation of BubR1 by Bub3 sustains mitotic PNAS PLUS checkpoint signaling Joo Seok Hana,b,1, Benjamin Vitrea,b,2, Daniele Fachinettia,b,2, and Don W. Clevelanda,b,c,1 aLudwig Institute for Cancer Research and Departments of bCellular and Molecular Medicine and cMedicine, University of California, San Diego, La Jolla, CA 92093 Contributed by Don W. Cleveland, August 25, 2014 (sent for review May 6, 2014) The mitotic checkpoint (also known as the spindle assembly sites) (18), along with its associated Bub3, but not Mad2, accounts checkpoint) prevents premature anaphase onset through gen- for the inhibition of APC/CCdc20 both in vitro using purified com- eration of an inhibitor of the E3 ubiquitin ligase APC/C, whose ponents and in vivo after induced degradation of either BubR1 or ubiquitination of cyclin B and securin targets them for degrada- Mad2 (13). These latter in vivo efforts were interpreted to dem- tion. Combining in vitro reconstitution and cell-based assays, we onstrate that Mad2 can dissociate from an initial complex with now identify dual mechanisms through which Bub3 promotes mi- Cdc20 (or APC/CCdc20) in an activated conformation capable of totic checkpoint signaling. Bub3 enhances signaling at unattached catalyzing additional Bub3–BubR1–Cdc20 complexes. Thus, we kinetochores not only by facilitating binding of BubR1 but also by proposed that BubR1–Bub3–Cdc20 is the mitotic checkpoint in- enhancing Cdc20 recruitment to kinetochores mediated by BubR1’s hibitor that blocks APC/C-dependent ubiquitination of cyclin B internal Cdc20 binding site. Downstream of kinetochore-produced and securin, through dual catalytic steps, an initial one at kinet- complexes, Bub3 promotes binding of BubR1’s conserved, amino ter- ochores and another acting within the cytoplasm mediated by minal Cdc20 binding domain to a site in Cdc20 that becomes exposed kinetochore-derived, activated Mad2 (13).