Ubiquitination of Cdc20 by the APC Occurs Through an Intramolecular Mechanism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A New Genetic Method for Isolating Functionally Interacting Genes
Copyright 2000 by the Genetics Society of America A New Genetic Method for Isolating Functionally Interacting Genes: High plo1؉-Dependent Mutants and Their Suppressors De®ne Genes in Mitotic and Septation Pathways in Fission Yeast C. Fiona Cullen,*,² Karen M. May,* Iain M. Hagan,³ David M. Glover²,§ and Hiroyuki Ohkura*,² *Institute of Cell and Molecular Biology, The University of Edinburgh, Edinburgh EH9 3JR, United Kingdom, ²Department of Anatomy and Physiology, Medical Sciences Institute, The University of Dundee, Dundee DD1 4HN, United Kingdom, ³School of Biological Sciences, The University of Manchester, Manchester M13 9PT, United Kingdom and §Department of Genetics, University of Cambridge, Cambridge CB2 3EH, United Kingdom Manuscript received February 2, 2000 Accepted for publication April 10, 2000 ABSTRACT We describe a general genetic method to identify genes encoding proteins that functionally interact with and/or are good candidates for downstream targets of a particular gene product. The screen identi®es mutants whose growth depends on high levels of expression of that gene. We apply this to the plo1ϩ gene that encodes a ®ssion yeast homologue of the polo-like kinases. plo1ϩ regulates both spindle formation and septation. We have isolated 17 high plo1ϩ-dependent (pld) mutants that show defects in mitosis or septation. Three mutants show a mitotic arrest phenotype. Among the 14 pld mutants with septation defects, 12 mapped to known loci: cdc7, cdc15, cdc11 spg1, and sid2. One of the pld mutants, cdc7-PD1, was selected for suppressor analysis. As multicopy suppressors, we isolated four known genes involved in septation in ®ssion yeast: spg1ϩ, sce3ϩ, cdc8ϩ, and rho1ϩ, and two previously uncharacterized genes, mpd1ϩ and mpd2ϩ. -

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

New Approaches to Functional Process Discovery in HPV 16-Associated Cervical Cancer Cells by Gene Ontology
Cancer Research and Treatment 2003;35(4):304-313 New Approaches to Functional Process Discovery in HPV 16-Associated Cervical Cancer Cells by Gene Ontology Yong-Wan Kim, Ph.D.1, Min-Je Suh, M.S.1, Jin-Sik Bae, M.S.1, Su Mi Bae, M.S.1, Joo Hee Yoon, M.D.2, Soo Young Hur, M.D.2, Jae Hoon Kim, M.D.2, Duck Young Ro, M.D.2, Joon Mo Lee, M.D.2, Sung Eun Namkoong, M.D.2, Chong Kook Kim, Ph.D.3 and Woong Shick Ahn, M.D.2 1Catholic Research Institutes of Medical Science, 2Department of Obstetrics and Gynecology, College of Medicine, The Catholic University of Korea, Seoul; 3College of Pharmacy, Seoul National University, Seoul, Korea Purpose: This study utilized both mRNA differential significant genes of unknown function affected by the display and the Gene Ontology (GO) analysis to char- HPV-16-derived pathway. The GO analysis suggested that acterize the multiple interactions of a number of genes the cervical cancer cells underwent repression of the with gene expression profiles involved in the HPV-16- cancer-specific cell adhesive properties. Also, genes induced cervical carcinogenesis. belonging to DNA metabolism, such as DNA repair and Materials and Methods: mRNA differential displays, replication, were strongly down-regulated, whereas sig- with HPV-16 positive cervical cancer cell line (SiHa), and nificant increases were shown in the protein degradation normal human keratinocyte cell line (HaCaT) as a con- and synthesis. trol, were used. Each human gene has several biological Conclusion: The GO analysis can overcome the com- functions in the Gene Ontology; therefore, several func- plexity of the gene expression profile of the HPV-16- tions of each gene were chosen to establish a powerful associated pathway, identify several cancer-specific cel- cervical carcinogenesis pathway. -

The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression
International Journal of Molecular Sciences Review The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression Tingting Zou and Zhenghong Lin * School of Life Sciences, Chongqing University, Chongqing 401331, China; [email protected] * Correspondence: [email protected] Abstract: The cell cycle is a collection of events by which cellular components such as genetic materials and cytoplasmic components are accurately divided into two daughter cells. The cell cycle transition is primarily driven by the activation of cyclin-dependent kinases (CDKs), which activities are regulated by the ubiquitin-mediated proteolysis of key regulators such as cyclins, CDK inhibitors (CKIs), other kinases and phosphatases. Thus, the ubiquitin-proteasome system (UPS) plays a pivotal role in the regulation of the cell cycle progression via recognition, interaction, and ubiquitination or deubiquitination of key proteins. The illegitimate degradation of tumor suppressor or abnormally high accumulation of oncoproteins often results in deregulation of cell proliferation, genomic instability, and cancer occurrence. In this review, we demonstrate the diversity and complexity of the regulation of UPS machinery of the cell cycle. A profound understanding of the ubiquitination machinery will provide new insights into the regulation of the cell cycle transition, cancer treatment, and the development of anti-cancer drugs. Keywords: cell cycle regulation; CDKs; cyclins; CKIs; UPS; E3 ubiquitin ligases; Deubiquitinases (DUBs) Citation: Zou, T.; Lin, Z. The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression. 1. Introduction Int. J. Mol. Sci. 2021, 22, 5754. https://doi.org/10.3390/ijms22115754 The cell cycle is a ubiquitous, complex, and highly regulated process that is involved in the sequential events during which a cell duplicates its genetic materials, grows, and di- Academic Editors: Kwang-Hyun Bae vides into two daughter cells. -

The APC/C in Female Mammalian Meiosis I
REPRODUCTIONREVIEW The APC/C in female mammalian meiosis I Hayden Homer1,2 1Mammalian Oocyte and Embryo Research Laboratory, Cell and Developmental Biology, UCL, London WC1E 6BT, UK and 2Reproductive Medicine Unit, Institute for Women’s Health, UCLH Elizabeth Garrett Anderson Wing, London NW1 2BU, UK Correspondence should be addressed to H Homer; Email: [email protected] Abstract The anaphase-promoting complex or cyclosome (APC/C) orchestrates a meticulously controlled sequence of proteolytic events critical for proper cell cycle progression, the details of which have been most extensively elucidated during mitosis. It has become apparent, however, that the APC/C, particularly when acting in concert with its Cdh1 co-activator (APC/CCdh1), executes a staggeringly diverse repertoire of functions that extend its remit well outside the bounds of mitosis. Findings over the past decade have not only earmarked mammalian oocyte maturation as one such case in point but have also begun to reveal a complex pattern of APC/C regulation that underpins many of the oocyte’s unique developmental attributes. This review will encompass the latest findings pertinent to the APC/C, especially APC/CCdh1, in mammalian oocytes and how its activity and substrates shape the stop–start tempo of female mammalian first meiotic division and the challenging requirement for assembling spindles in the absence of centrosomes. Reproduction (2013) 146 R61–R71 Introduction associated somatic follicular compartment at the time of ovulation. Significantly, although primordial germ cells Meiosis is the unique cell division that halves the in the ovary commit to meiosis during fetal life, it is chromosome compliment by coupling two successive not until postnatal adulthood that mature oocytes (or nuclear divisions with a single round of DNA replication. -

1 Spindle Assembly Checkpoint Is Sufficient for Complete Cdc20
Spindle assembly checkpoint is sufficient for complete Cdc20 sequestering in mitotic control Bashar Ibrahim Bio System Analysis Group, Friedrich-Schiller-University Jena, and Jena Centre for Bioinformatics (JCB), 07743 Jena, Germany Email: [email protected] Abstract The spindle checkpoint assembly (SAC) ensures genome fidelity by temporarily delaying anaphase onset, until all chromosomes are properly attached to the mitotic spindle. The SAC delays mitotic progression by preventing activation of the ubiquitin ligase anaphase-promoting complex (APC/C) or cyclosome; whose activation by Cdc20 is required for sister-chromatid separation marking the transition into anaphase. The mitotic checkpoint complex (MCC), which contains Cdc20 as a subunit, binds stably to the APC/C. Compelling evidence by Izawa and Pines (Nature 2014; 10.1038/nature13911) indicates that the MCC can inhibit a second Cdc20 that has already bound and activated the APC/C. Whether or not MCC per se is sufficient to fully sequester Cdc20 and inhibit APC/C remains unclear. Here, a dynamic model for SAC regulation in which the MCC binds a second Cdc20 was constructed. This model is compared to the MCC, and the MCC-and-BubR1 (dual inhibition of APC) core model variants and subsequently validated with experimental data from the literature. By using ordinary nonlinear differential equations and spatial simulations, it is shown that the SAC works sufficiently to fully sequester Cdc20 and completely inhibit APC/C activity. This study highlights the principle that a systems biology approach is vital for molecular biology and could also be used for creating hypotheses to design future experiments. Keywords: Mathematical biology, Spindle assembly checkpoint; anaphase promoting complex, MCC, Cdc20, systems biology 1 Introduction Faithful DNA segregation, prior to cell division at mitosis, is vital for maintaining genomic integrity. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Bub1 Positions Mad1 Close to KNL1 MELT Repeats to Promote Checkpoint Signalling
ARTICLE Received 14 Dec 2016 | Accepted 3 May 2017 | Published 12 June 2017 DOI: 10.1038/ncomms15822 OPEN Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling Gang Zhang1, Thomas Kruse1, Blanca Lo´pez-Me´ndez1, Kathrine Beck Sylvestersen1, Dimitriya H. Garvanska1, Simone Schopper1, Michael Lund Nielsen1 & Jakob Nilsson1 Proper segregation of chromosomes depends on a functional spindle assembly checkpoint (SAC) and requires kinetochore localization of the Bub1 and Mad1/Mad2 checkpoint proteins. Several aspects of Mad1/Mad2 kinetochore recruitment in human cells are unclear and in particular the underlying direct interactions. Here we show that conserved domain 1 (CD1) in human Bub1 binds directly to Mad1 and a phosphorylation site exists in CD1 that stimulates Mad1 binding and SAC signalling. Importantly, fusion of minimal kinetochore-targeting Bub1 fragments to Mad1 bypasses the need for CD1, revealing that the main function of Bub1 is to position Mad1 close to KNL1 MELTrepeats. Furthermore, we identify residues in Mad1 that are critical for Mad1 functionality, but not Bub1 binding, arguing for a direct role of Mad1 in the checkpoint. This work dissects functionally relevant molecular interactions required for spindle assembly checkpoint signalling at kinetochores in human cells. 1 The Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3B, 2200 Copenhagen, Denmark. Correspondence and requests for materials should be addressed to G.Z. -

A 20S Complex Containing CDC27 and CDC16 Catalyzes the Mitosis-Specific Conjugation of Ubiquitin to Cyclin B
Cell, Vol. 81,279-288, April 21, 1995, Copyright© 1995 by Cell Press A 20S Complex Containing CDC27 and CDC16 Catalyzes the Mitosis-Specific Conjugation of Ubiquitin to Cyclin B Randall W. King,*t Jan-Michael Peters,*t cyclin degradation is required to exit mitosis (Surana et Stuart Tugendreich,$ Mark Rolfe,§ Philip Hieter,$ al., 1993; Holloway et al., 1993). Treatments that interfere and Marc W. Kirschnert with the proteolysis of endogenous cyclin B, however, ar- tDepartment of Cell Biology rest cell division earlier, at anaphase (Holloway et al., Harvard Medical School 1993). This discrepancy can be explained by hypothesiz- Boston, Massachusetts 02115 ing that chromosome segregation and cyclin proteolysis $Department of Molecular Biology and Genetics depend upon common components. Support for this idea Johns Hopkins School of Medicine has emerged recently from studies in budding yeast, in Baltimore, Maryland 21205 which CDC16 and CDC23, genes required for progression §Mitotix, Incorporated through anaphase, have been shown to be required for One Kendall Square the proteolysis of B-type cyclins (Irniger et al., 1995 [this Cambridge, Massachusetts 02139 issue of Cell]). The proteins encoded by these genes form a complex with the CDC27 protein (Lamb et al., 1994), a homolog of which is also required for anaphase progres- Summary sion in mammalian cells (Tugendreich et al., 1995 [this issue of Cell]). Cyclin B is degraded at the onset of anaphase by a Biochemical evidence suggests that cyclin proteolysis ubiquitin-dependent proteolytic system. We have frac- is mediated by the ubiquitin pathway: cyclin B-ubiquitin tionated mitotic Xenopus egg extracts to identify com- conjugates can be observed in mitotic, but not interphase, ponents required for this process. -

Patronus Is the Elusive Plant Securin, Preventing Chromosome Separation By
bioRxiv preprint doi: https://doi.org/10.1101/606285; this version posted April 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Patronus is the elusive plant securin, preventing chromosome separation by 2 antagonizing separase 3 Laurence Cromer1, Sylvie Jolivet1, Dipesh Kumar Singh1, Floriane Berthier1, Nancy 4 De Winne2, Geert De Jaeger2, Shinichiro Komaki3,4, Maria Ada Prusicki3, Arp 5 Schnittger3, Raphael Guérois5 and Raphael Mercier1* 6 7 1 Institut Jean-Pierre Bourgin, UMR1318 INRA-AgroParisTech, Université Paris- 8 Saclay, RD10, 78000 Versailles, France. 9 2 Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, 10 Belgium, VIB Center for Plant Systems Biology, Ghent, Belgium 11 3 Department of Developmental Biology, Institute for Plant Sciences and 12 Microbiology, University of Hamburg, 22609 Hamburg, Germany. 13 4 Present address : Nara Institute of Science and Technology, Graduate School of 14 Biological Sciences, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan 15 5 Institute for Integrative Biology of the Cell (I2BC), Commissariat à l’Energie 16 Atomique et aux Energies Alternatives (CEA), Centre National de la Recherche 17 Scientifique (CNRS), Université Paris-Sud, CEA-Saclay, Gif-sur-Yvette, France 18 * Corresponding author. [email protected] 19 1 bioRxiv preprint doi: https://doi.org/10.1101/606285; this version posted April 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Protein Engineering of a Ubiquitin-Variant Inhibitor of APC/C Identifies a Cryptic K48 Ubiquitin Chain Binding Site
Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site Edmond R. Watsona,b, Christy R. R. Gracea, Wei Zhangc,d,2, Darcie J. Millera, Iain F. Davidsone, J. Rajan Prabub, Shanshan Yua, Derek L. Bolhuisf, Elizaveta T. Kulkof, Ronnald Vollrathb, David Haselbache,g, Holger Starkg, Jan-Michael Peterse,h, Nicholas G. Browna,f, Sachdev S. Sidhuc,1, and Brenda A. Schulmana,b,1 aDepartment of Structural Biology, St. Jude Children’s Research Hospital, Memphis, TN 38105; bDepartment of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, 82152 Martinsried, Germany; cDonnelly Centre for Cellular and Biomolecular Research, Banting and Best Department of Medical Research, University of Toronto, Toronto, ON, Canada M5S3E1; dDepartment of Molecular Genetics, University of Toronto, Toronto, ON, Canada M5S3E1; eResearch Institute of Molecular Pathology, Vienna BioCenter, 1030 Vienna, Austria; fDepartment of Pharmacology and Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, NC 27599; gMax Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany; and hMedical University of Vienna, 1090 Vienna, Austria Contributed by Brenda A. Schulman, June 24, 2019 (sent for review February 19, 2019; reviewed by Kylie Walters and Hao Wu) Ubiquitin (Ub)-mediated proteolysis is a fundamental mechanism the type of catalytic domain. E3s harboring “HECT” and “RBR” used by eukaryotic cells to maintain homeostasis and protein catalytic domains promote ubiquitylation through 2-step reac- quality, and to control timing in biological processes. Two essential tions involving formation of a thioester-linked intermediate be- aspects of Ub regulation are conjugation through E1-E2-E3 enzy- tween the E3 and Ub’s C terminus: First, Ub is transferred from matic cascades and recognition by Ub-binding domains. -
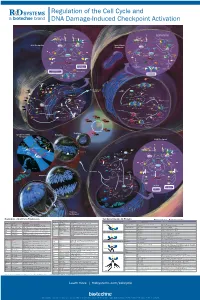
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor