Mitotic Cdks Control the Metaphase–Anaphase Transition and Trigger Spindle Elongation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Meiotic Prophase Abnormalities and Metaphase Cell Death in MLH1-Deficient Mouse Spermatocytes: Insights Into Regulation of Spermatogenic Progress
Developmental Biology 249, 85–95 (2002) doi:10.1006/dbio.2002.0708 Meiotic Prophase Abnormalities and Metaphase Cell Death in MLH1-Deficient Mouse Spermatocytes: Insights into Regulation of Spermatogenic Progress Shannon Eaker,1 John Cobb,2 April Pyle, and Mary Ann Handel3 Department of Biochemistry and Cellular and Molecular Biology, University of Tennessee, Knoxville, Tennessee 37996 The MLH1 protein is required for normal meiosis in mice and its absence leads to failure in maintenance of pairing between bivalent chromosomes, abnormal meiotic division, and ensuing sterility in both sexes. In this study, we investigated whether failure to develop foci of MLH1 protein on chromosomes in prophase would lead to elimination of prophase spermatocytes, and, if not, whether univalent chromosomes could align normally on the meiotic spindle and whether metaphase spermatocytes would be delayed and/or eliminated. In spite of the absence of MLH1 foci, no apoptosis of spermatocytes in prophase was detected. In fact, chromosomes of pachytene spermatocytes from Mlh1؊/؊ mice were competent to condense metaphase chromosomes, both in vivo and in vitro. Most condensed chromosomes were univalents with spatially distinct FISH signals. Typical metaphase events, such as synaptonemal complex breakdown and the phosphorylation of Ser10 on histone H3, occurred in Mlh1؊/؊ spermatocytes, suggesting that there is no inhibition of onset of meiotic metaphase in the face of massive chromosomal abnormalities. However, the condensed univalent chromosomes did not align correctly onto the spindle apparatus in the majority of Mlh1؊/؊ spermatocytes. Most meiotic metaphase spermatocytes were characterized with bipolar spindles, but chromosomes radiated away from the microtubule-organizing centers in a prometaphase-like pattern rather than achieving a bipolar orientation. -

Mitosis Vs. Meiosis
Mitosis vs. Meiosis In order for organisms to continue growing and/or replace cells that are dead or beyond repair, cells must replicate, or make identical copies of themselves. In order to do this and maintain the proper number of chromosomes, the cells of eukaryotes must undergo mitosis to divide up their DNA. The dividing of the DNA ensures that both the “old” cell (parent cell) and the “new” cells (daughter cells) have the same genetic makeup and both will be diploid, or containing the same number of chromosomes as the parent cell. For reproduction of an organism to occur, the original parent cell will undergo Meiosis to create 4 new daughter cells with a slightly different genetic makeup in order to ensure genetic diversity when fertilization occurs. The four daughter cells will be haploid, or containing half the number of chromosomes as the parent cell. The difference between the two processes is that mitosis occurs in non-reproductive cells, or somatic cells, and meiosis occurs in the cells that participate in sexual reproduction, or germ cells. The Somatic Cell Cycle (Mitosis) The somatic cell cycle consists of 3 phases: interphase, m phase, and cytokinesis. 1. Interphase: Interphase is considered the non-dividing phase of the cell cycle. It is not a part of the actual process of mitosis, but it readies the cell for mitosis. It is made up of 3 sub-phases: • G1 Phase: In G1, the cell is growing. In most organisms, the majority of the cell’s life span is spent in G1. • S Phase: In each human somatic cell, there are 23 pairs of chromosomes; one chromosome comes from the mother and one comes from the father. -

Cell Division- Ch 5
Cell Division- Mitosis and Meiosis When do cells divide? Cell size . One of most important factors affecting size of the cell is size of cell membrane . Cell must remain relatively small to survive (why?) – Cell membrane has to be big enough to take in nutrients and eliminate wastes – As cells get bigger, the volume increases faster than the surface area – Small cells have a larger surface area to volume ratio than larger cells to help with nutrient intake and waste elimination . When a cell reaches its max size, the nucleus starts cell division: called MITOSIS or MEIOSIS Mitosis . General Information – Occurs in somatic (body) cells ONLY!! – Nickname: called “normal” cell division – Produces somatic cells only . Background Info – Starts with somatic cell in DIPLOID (2n) state . Cell contains homologous chromosomes- chromosomes that control the same traits but not necessarily in the same way . 1 set from mom and 1 set from dad – Ends in diploid (2n) state as SOMATIC cells – Goes through one set of divisions – Start with 1 cell and end with 2 cells Mitosis (cont.) . Accounts for three essential life processes – Growth . Result of cell producing new cells . Develop specialized shapes/functions in a process called differentiation . Rate of cell division controlled by GH (Growth Hormone) which is produced in the pituitary gland . Ex. Nerve cell, intestinal cell, etc. – Repair . Cell regenerates at the site of injury . Ex. Skin (replaced every 28 days), blood vessels, bone Mitosis (cont.) – Reproduction . Asexual – Offspring produced by only one parent – Produce offspring that are genetically identical – MITOSIS – Ex. Bacteria, fungi, certain plants and animals . -

List, Describe, Diagram, and Identify the Stages of Meiosis
Meiosis and Sexual Life Cycles Objective # 1 In this topic we will examine a second type of cell division used by eukaryotic List, describe, diagram, and cells: meiosis. identify the stages of meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis and meiosis, are involved in transmitting genetic information from one generation to the next during eukaryotic life cycles. 1 2 Objective 1 Objective 1 Overview of meiosis in a cell where 2N = 6 Only diploid cells can divide by meiosis. We will examine the stages of meiosis in DNA duplication a diploid cell where 2N = 6 during interphase Meiosis involves 2 consecutive cell divisions. Since the DNA is duplicated Meiosis II only prior to the first division, the final result is 4 haploid cells: Meiosis I 3 After meiosis I the cells are haploid. 4 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis Prophase I: ¾ Chromosomes condense. Because of replication during interphase, each chromosome consists of 2 sister chromatids joined by a centromere. ¾ Synapsis – the 2 members of each homologous pair of chromosomes line up side-by-side to form a tetrad consisting of 4 chromatids: 5 6 1 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis Prophase I: ¾ During synapsis, sometimes there is an exchange of homologous parts between non-sister chromatids. This exchange is called crossing over. 7 8 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis (2N=6) Prophase I: ¾ the spindle apparatus begins to form. ¾ the nuclear membrane breaks down: Prophase I 9 10 Objective 1, Stages of Meiosis Objective 1, 4 Possible Metaphase I Arrangements: Metaphase I: ¾ chromosomes line up along the equatorial plate in pairs, i.e. -

Opium Alkaloid Noscapine Is an Antitumor Agent That Arrests Metaphase and Induces Apoptosis in Dividing Cells
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 1601–1606, February 1998 Cell Biology Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells KEQIANG YE*†,YONG KE‡,NAGALAKSHMI KESHAVA§,JOHN SHANKS†,JUDITH A. KAPP‡,RAJESHWAR R. TEKMAL§, i JOHN PETROS¶, AND HARISH C. JOSHI*† *Graduate Program in Biochemistry and Molecular Biology, Departments of †Anatomy and Cell Biology, ‡Pathology, §Gynecology–Obstetrics, and ¶Urology, Emory University School of Medicine, Atlanta, GA 30322 Edited by Thomas D. Pollard, Salk Institute for Biological Studies, La Jolla, CA, and approved December 17, 1997 (received for review July 17, 1997) ABSTRACT An alkaloid from opium, noscapine, is used (2,3,4-trimethoxyphenyl)-2,4,6-cycloheptatrien-1-one], TCB as an antitussive drug and has low toxicity in humans and (2,3,4-trimethoxy-49-carbomethoxy-1,19-biphenyl), and TKB mice. We show that noscapine binds stoichiometrically to (2,3,4-trimethoxy-49-acetyl-1,19-biphenyl), and vinca alkaloids. tubulin, alters its conformation, affects microtubule assembly, Taxol and its analogs represent the compounds that promote and arrests mammalian cells in mitosis. Furthermore, the assembly of microtubules. It is now clear that although all noscapine causes apoptosis in many cell types and has potent of these microtubule drugs prevent cell division, only a select antitumor activity against solid murine lymphoid tumors few have been useful clinically. In addition, there are differ- (even when the drug was administered orally) and against ences regarding the toxicity and the efficacy of these drugs for human breast and bladder tumors implanted in nude mice. -

Cell Life Cycle and Reproduction the Cell Cycle (Cell-Division Cycle), Is a Series of Events That Take Place in a Cell Leading to Its Division and Duplication
Cell Life Cycle and Reproduction The cell cycle (cell-division cycle), is a series of events that take place in a cell leading to its division and duplication. The main phases of the cell cycle are interphase, nuclear division, and cytokinesis. Cell division produces two daughter cells. In cells without a nucleus (prokaryotic), the cell cycle occurs via binary fission. Interphase Gap1(G1)- Cells increase in size. The G1checkpointcontrol mechanism ensures that everything is ready for DNA synthesis. Synthesis(S)- DNA replication occurs during this phase. DNA Replication The process in which DNA makes a duplicate copy of itself. Semiconservative Replication The process in which the DNA molecule uncoils and separates into two strands. Each original strand becomes a template on which a new strand is constructed, resulting in two DNA molecules identical to the original DNA molecule. Gap 2(G2)- The cell continues to grow. The G2checkpointcontrol mechanism ensures that everything is ready to enter the M (mitosis) phase and divide. Mitotic(M) refers to the division of the nucleus. Cell growth stops at this stage and cellular energy is focused on the orderly division into daughter cells. A checkpoint in the middle of mitosis (Metaphase Checkpoint) ensures that the cell is ready to complete cell division. The final event is cytokinesis, in which the cytoplasm divides and the single parent cell splits into two daughter cells. Reproduction Cellular reproduction is a process by which cells duplicate their contents and then divide to yield multiple cells with similar, if not duplicate, contents. Mitosis Mitosis- nuclear division resulting in the production of two somatic cells having the same genetic complement (genetically identical) as the original cell. -

Patronus Is the Elusive Plant Securin, Preventing Chromosome Separation By
bioRxiv preprint doi: https://doi.org/10.1101/606285; this version posted April 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Patronus is the elusive plant securin, preventing chromosome separation by 2 antagonizing separase 3 Laurence Cromer1, Sylvie Jolivet1, Dipesh Kumar Singh1, Floriane Berthier1, Nancy 4 De Winne2, Geert De Jaeger2, Shinichiro Komaki3,4, Maria Ada Prusicki3, Arp 5 Schnittger3, Raphael Guérois5 and Raphael Mercier1* 6 7 1 Institut Jean-Pierre Bourgin, UMR1318 INRA-AgroParisTech, Université Paris- 8 Saclay, RD10, 78000 Versailles, France. 9 2 Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, 10 Belgium, VIB Center for Plant Systems Biology, Ghent, Belgium 11 3 Department of Developmental Biology, Institute for Plant Sciences and 12 Microbiology, University of Hamburg, 22609 Hamburg, Germany. 13 4 Present address : Nara Institute of Science and Technology, Graduate School of 14 Biological Sciences, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan 15 5 Institute for Integrative Biology of the Cell (I2BC), Commissariat à l’Energie 16 Atomique et aux Energies Alternatives (CEA), Centre National de la Recherche 17 Scientifique (CNRS), Université Paris-Sud, CEA-Saclay, Gif-sur-Yvette, France 18 * Corresponding author. [email protected] 19 1 bioRxiv preprint doi: https://doi.org/10.1101/606285; this version posted April 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Review Questions Meiosis
Review Questions Meiosis 1. Asexual reproduction versus sexual reproduction: which is better? Asexual reproduction is much more efficient than sexual reproduction in a number of ways. An organism doesn’t have to find a mate. An organism donates 100% of its’ genetic material to its offspring (with sex, only 50% end up in the offspring). All members of a population can produce offspring, not just females, enabling asexual organisms to out-reproduce sexual rivals. 2. So why is there sex? Why are there boys? If females can reproduce easier and more efficiently asexually, then why bother with males? Sex is good for evolution because it creates genetic variety. All organisms depend on mutations for genetic variation. Sex takes these preexisting traits (created by mutations) and shuffles them into new combinations (genetic recombination). For example, if we wanted a rice plant that was fast-growing but also had a high yield, we would have to wait a long time for a fast-growing rice to undergo a mutation that would also make it highly productive. An easy way to combine these two desirable traits is through sexually reproduction. By breeding a fast-growing variety with a high-yielding variety, we can create offspring with both traits. In an asexual organism, all the offspring are genetically identical to the parent (unless there was a mutation) and genetically identically to each other. Sexual reproduction creates offspring that are genetically different from the parents and genetically different from their siblings. In a stable environment, asexual reproduction may work just fine. However, most ecosystems are dynamic places. -

Regulation of Cyclin-Dependent Kinase Activity During Mitotic Exit and Maintenance of Genome Stability by P21, P27, and P107
Regulation of cyclin-dependent kinase activity during mitotic exit and maintenance of genome stability by p21, p27, and p107 Taku Chibazakura*†, Seth G. McGrew‡§, Jonathan A. Cooper§, Hirofumi Yoshikawa*, and James M. Roberts‡§ *Deparment of Bioscience, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya-ku, Tokyo 156-8502, Japan; and ‡Howard Hughes Medical Institute and §Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA 98019 Communicated by Robert N. Eisenman, Fred Hutchinson Cancer Research Center, Seattle, WA, February 4, 2004 (received for review October 28, 2003) To study the regulation of cyclin-dependent kinase (CDK) activity bind to and inactivate mitotic cyclin–CDK complexes (15, 16). during mitotic exit in mammalian cells, we constructed murine cell These CKIs accumulate and persist during mid-M-to-G1 phase ͞ lines that constitutively express a stabilized mutant of cyclin A until they are phosphorylated by Sic1 Rum1-resistant G1 cyclin- (cyclin A47). Even though cyclin A47 was expressed throughout CDKs, which initiates their ubiquitin-dependent degradation at mitosis and in G1 cells, its associated CDK activity was inactivated the G1-to-S phase transition (17–19). Thus, Sic1 and Rum1 after the transition from metaphase to anaphase. Cyclin A47 constitute a switch that controls the transition from a state of low associated with both p21 and p27 during mitotic exit, implicating CDK activity to that of high CDK activity, thereby regulating these proteins in CDK inactivation. However, cyclin A47 was fully mitotic exit and S phase entry. This parallels the activity of ؊/؊ ؊/؊ inhibited during the M-to-G1 transition in p21 p27 fibro- APC-Cdh1, and indeed these two pathways constitute redundant blasts. -
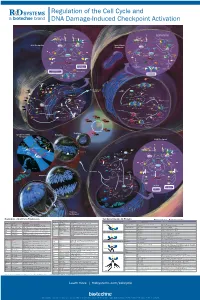
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

Control of the Cell Cycle During Meiotic Maturation of Mouse Oocytes
Control of the cell cycle during meiotic maturation of mouse oocytes Ibtissem Nabti Department of Cell and Developmental Biology University College London London, UK A thesis submitted for the degree of Doctor of Philosophy April 2010 Declaration The work presented in this thesis was conducted under the supervision of Professor John Carroll in the department of cell and developmental biology, University College London, and Professor Keith Jones in the department of reproductive physiology, University of Newcastle upon Tyne. I, Ibtissem Nabti confirm that the data presented in this thesis is my own and from original studies. Also, this dissertation has not previously been submitted, wholly or in part, to any other university for any degree or diploma. Ibtissem Nabti 1 Abstract The overall aim of the work presented in this thesis is to better understand the molecular mechanisms underlying the progression of the meiotic division in mammalian oocytes. The first goal of this project was to investigate the relative contribution of securin and cyclin B1 to the control of the protease separase during the indeterminate period of metaphase II arrest. I found here that although there are conditions in which either securin or MPF can prevent chromosome disjunction, such as during meiosis I, securin is the predominant inhibitor of separase during metaphase II arrest. Thus, CDK1 pharmacological inhibition as well as antibody inhibition of CDK1/cyclin B1 binding to separase, both failed to induce sister chromatid disjunction. Instead, securin morpholino knockdown induced sister chromatid separation, which could be rescued by injection of securin cRNA. I also examined the effect of CDK1 and MAPK activities on securin stability. -

Meiosis I and Meiosis II; Life Cycles
Meiosis I and Meiosis II; Life Cycles Meiosis functions to reduce the number of chromosomes to one half. Each daughter cell that is produced will have one half as many chromosomes as the parent cell. Meiosis is part of the sexual process because gametes (sperm, eggs) have one half the chromosomes as diploid (2N) individuals. Phases of Meiosis There are two divisions in meiosis; the first division is meiosis I: the number of cells is doubled but the number of chromosomes is not. This results in 1/2 as many chromosomes per cell. The second division is meiosis II: this division is like mitosis; the number of chromosomes does not get reduced. The phases have the same names as those of mitosis. Meiosis I: prophase I (2N), metaphase I (2N), anaphase I (N+N), and telophase I (N+N) Meiosis II: prophase II (N+N), metaphase II (N+N), anaphase II (N+N+N+N), and telophase II (N+N+N+N) (Works Cited See) *3 Meiosis I (Works Cited See) *1 1. Prophase I Events that occur during prophase of mitosis also occur during prophase I of meiosis. The chromosomes coil up, the nuclear membrane begins to disintegrate, and the centrosomes begin moving apart. The two chromosomes may exchange fragments by a process called crossing over. When the chromosomes partially separate in late prophase, until they separate during anaphase resulting in chromosomes that are mixtures of the original two chromosomes. 2. Metaphase I Bivalents (tetrads) become aligned in the center of the cell and are attached to spindle fibers.