Structural Analysis of Bub3 Interactions in the Mitotic Spindle Checkpoint
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

UNIVERSITY of CALIFORNIA, SAN DIEGO Functional Analysis of Sall4
UNIVERSITY OF CALIFORNIA, SAN DIEGO Functional analysis of Sall4 in modulating embryonic stem cell fate A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Molecular Pathology by Pei Jen A. Lee Committee in charge: Professor Steven Briggs, Chair Professor Geoff Rosenfeld, Co-Chair Professor Alexander Hoffmann Professor Randall Johnson Professor Mark Mercola 2009 Copyright Pei Jen A. Lee, 2009 All rights reserved. The dissertation of Pei Jen A. Lee is approved, and it is acceptable in quality and form for publication on microfilm and electronically: ______________________________________________________________ ______________________________________________________________ ______________________________________________________________ ______________________________________________________________ Co-Chair ______________________________________________________________ Chair University of California, San Diego 2009 iii Dedicated to my parents, my brother ,and my husband for their love and support iv Table of Contents Signature Page……………………………………………………………………….…iii Dedication…...…………………………………………………………………………..iv Table of Contents……………………………………………………………………….v List of Figures…………………………………………………………………………...vi List of Tables………………………………………………….………………………...ix Curriculum vitae…………………………………………………………………………x Acknowledgement………………………………………………….……….……..…...xi Abstract………………………………………………………………..…………….....xiii Chapter 1 Introduction ..…………………………………………………………………………….1 Chapter 2 Materials and Methods……………………………………………………………..…12 -

Viral Strategies to Arrest Host Mrna Nuclear Export
Viruses 2013, 5, 1824-1849; doi:10.3390/v5071824 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review Nuclear Imprisonment: Viral Strategies to Arrest Host mRNA Nuclear Export Sharon K. Kuss *, Miguel A. Mata, Liang Zhang and Beatriz M. A. Fontoura Department of Cell Biology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA; E-Mails: [email protected] (M.A.M.); [email protected] (L.Z.); [email protected] (B.M.A.F) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-214-633-2001; Fax: +1-214-648-5814. Received: 10 June 2013; in revised form: 27 June 2013 / Accepted: 11 July 2013 / Published: 18 July 2013 Abstract: Viruses possess many strategies to impair host cellular responses to infection. Nuclear export of host messenger RNAs (mRNA) that encode antiviral factors is critical for antiviral protein production and control of viral infections. Several viruses have evolved sophisticated strategies to inhibit nuclear export of host mRNAs, including targeting mRNA export factors and nucleoporins to compromise their roles in nucleo-cytoplasmic trafficking of cellular mRNA. Here, we present a review of research focused on suppression of host mRNA nuclear export by viruses, including influenza A virus and vesicular stomatitis virus, and the impact of this viral suppression on host antiviral responses. Keywords: virus; influenza virus; vesicular stomatitis virus; VSV; NS1; matrix protein; nuclear export; nucleo-cytoplasmic trafficking; mRNA export; NXF1; TAP; CRM1; Rae1 1. Introduction Nucleo-cytoplasmic trafficking of proteins and RNA is critical for proper cellular functions and survival. -

F-Box Protein RAE1 Regulates the Stability of the Aluminum-Resistance Transcription Factor STOP1 in Arabidopsis
F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis Yang Zhanga,b,1, Jie Zhanga,1, Jinliang Guoa,b,1, Fanglin Zhoua, Somesh Singha, Xuan Xub, Qi Xiec, Zhongbao Yangd, and Chao-Feng Huanga,b,2 aNational Key Laboratory of Plant Molecular Genetics, Shanghai Center for Plant Stress Biology, Center of Excellence for Molecular Plant Sciences, Chinese Academy of Sciences, 200032 Shanghai, China; bCollege of Resources and Environmental Sciences, Nanjing Agricultural University, 210095 Nanjing, China; cState Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, 100101 Beijing, China; and dKey Laboratory of Plant Cell Engineering and Germplasm Innovation, Ministry of Education, College of Life Science, Shandong University, 250100 Jinan, China Edited by Luis Herrera-Estrella, Center for Research and Advanced Studies, Irapuato, Mexico, and approved November 19, 2018 (received for review August 21, 2018) Aluminum (Al) toxicity is a major factor limiting crop production expression of STOP1-downstream Al-resistance genes, including on acid soils, which represent over 30% of the world’s arable land. AtALMT1, AtMATE, and ALS3, is induced by Al (8, 12). These Some plants have evolved mechanisms to detoxify Al. Arabidopsis, results suggest the possibility that STOP1 might be regulated by for example, secretes malate via the AtALMT1 transporter to che- Al at posttranscriptional or posttranslational levels. late and detoxify Al. The C2H2-type transcription factor STOP1 plays a Ubiquitin-mediated protein degradation is an important post- crucial role in Al resistance by inducing the expression of a set of translational mechanism that regulates numerous biological processes genes, including AtALMT1. -

1 Spindle Assembly Checkpoint Is Sufficient for Complete Cdc20
Spindle assembly checkpoint is sufficient for complete Cdc20 sequestering in mitotic control Bashar Ibrahim Bio System Analysis Group, Friedrich-Schiller-University Jena, and Jena Centre for Bioinformatics (JCB), 07743 Jena, Germany Email: [email protected] Abstract The spindle checkpoint assembly (SAC) ensures genome fidelity by temporarily delaying anaphase onset, until all chromosomes are properly attached to the mitotic spindle. The SAC delays mitotic progression by preventing activation of the ubiquitin ligase anaphase-promoting complex (APC/C) or cyclosome; whose activation by Cdc20 is required for sister-chromatid separation marking the transition into anaphase. The mitotic checkpoint complex (MCC), which contains Cdc20 as a subunit, binds stably to the APC/C. Compelling evidence by Izawa and Pines (Nature 2014; 10.1038/nature13911) indicates that the MCC can inhibit a second Cdc20 that has already bound and activated the APC/C. Whether or not MCC per se is sufficient to fully sequester Cdc20 and inhibit APC/C remains unclear. Here, a dynamic model for SAC regulation in which the MCC binds a second Cdc20 was constructed. This model is compared to the MCC, and the MCC-and-BubR1 (dual inhibition of APC) core model variants and subsequently validated with experimental data from the literature. By using ordinary nonlinear differential equations and spatial simulations, it is shown that the SAC works sufficiently to fully sequester Cdc20 and completely inhibit APC/C activity. This study highlights the principle that a systems biology approach is vital for molecular biology and could also be used for creating hypotheses to design future experiments. Keywords: Mathematical biology, Spindle assembly checkpoint; anaphase promoting complex, MCC, Cdc20, systems biology 1 Introduction Faithful DNA segregation, prior to cell division at mitosis, is vital for maintaining genomic integrity. -

BUB3 That Dissociates from BUB1 Activates Caspase-Independent Mitotic Death (CIMD)
Cell Death and Differentiation (2010) 17, 1011–1024 & 2010 Macmillan Publishers Limited All rights reserved 1350-9047/10 $32.00 www.nature.com/cdd BUB3 that dissociates from BUB1 activates caspase-independent mitotic death (CIMD) Y Niikura1, H Ogi1, K Kikuchi1 and K Kitagawa*,1 The cell death mechanism that prevents aneuploidy caused by a failure of the spindle checkpoint has recently emerged as an important regulatory paradigm. We previously identified a new type of mitotic cell death, termed caspase-independent mitotic death (CIMD), which is induced during early mitosis by partial BUB1 (a spindle checkpoint protein) depletion and defects in kinetochore–microtubule attachment. In this study, we have shown that survived cells that escape CIMD have abnormal nuclei, and we have determined the molecular mechanism by which BUB1 depletion activates CIMD. The BUB3 protein (a BUB1 interactor and a spindle checkpoint protein) interacts with p73 (a homolog of p53), specifically in cells wherein CIMD occurs. The BUB3 protein that is freed from BUB1 associates with p73 on which Y99 is phosphorylated by c-Abl tyrosine kinase, resulting in the activation of CIMD. These results strongly support the hypothesis that CIMD is the cell death mechanism protecting cells from aneuploidy by inducing the death of cells prone to substantial chromosome missegregation. Cell Death and Differentiation (2010) 17, 1011–1024; doi:10.1038/cdd.2009.207; published online 8 January 2010 Aneuploidy – the presence of an abnormal number of of spindle checkpoint activity.20,21 -

The Genome of Schmidtea Mediterranea and the Evolution Of
OPEN ArtICLE doi:10.1038/nature25473 The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms Markus Alexander Grohme1*, Siegfried Schloissnig2*, Andrei Rozanski1, Martin Pippel2, George Robert Young3, Sylke Winkler1, Holger Brandl1, Ian Henry1, Andreas Dahl4, Sean Powell2, Michael Hiller1,5, Eugene Myers1 & Jochen Christian Rink1 The planarian Schmidtea mediterranea is an important model for stem cell research and regeneration, but adequate genome resources for this species have been lacking. Here we report a highly contiguous genome assembly of S. mediterranea, using long-read sequencing and a de novo assembler (MARVEL) enhanced for low-complexity reads. The S. mediterranea genome is highly polymorphic and repetitive, and harbours a novel class of giant retroelements. Furthermore, the genome assembly lacks a number of highly conserved genes, including critical components of the mitotic spindle assembly checkpoint, but planarians maintain checkpoint function. Our genome assembly provides a key model system resource that will be useful for studying regeneration and the evolutionary plasticity of core cell biological mechanisms. Rapid regeneration from tiny pieces of tissue makes planarians a prime De novo long read assembly of the planarian genome model system for regeneration. Abundant adult pluripotent stem cells, In preparation for genome sequencing, we inbred the sexual strain termed neoblasts, power regeneration and the continuous turnover of S. mediterranea (Fig. 1a) for more than 17 successive sib- mating of all cell types1–3, and transplantation of a single neoblast can rescue generations in the hope of decreasing heterozygosity. We also developed a lethally irradiated animal4. Planarians therefore also constitute a a new DNA isolation protocol that meets the purity and high molecular prime model system for stem cell pluripotency and its evolutionary weight requirements of PacBio long-read sequencing12 (Extended Data underpinnings5. -

Patronus Is the Elusive Plant Securin, Preventing Chromosome Separation By
bioRxiv preprint doi: https://doi.org/10.1101/606285; this version posted April 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Patronus is the elusive plant securin, preventing chromosome separation by 2 antagonizing separase 3 Laurence Cromer1, Sylvie Jolivet1, Dipesh Kumar Singh1, Floriane Berthier1, Nancy 4 De Winne2, Geert De Jaeger2, Shinichiro Komaki3,4, Maria Ada Prusicki3, Arp 5 Schnittger3, Raphael Guérois5 and Raphael Mercier1* 6 7 1 Institut Jean-Pierre Bourgin, UMR1318 INRA-AgroParisTech, Université Paris- 8 Saclay, RD10, 78000 Versailles, France. 9 2 Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, 10 Belgium, VIB Center for Plant Systems Biology, Ghent, Belgium 11 3 Department of Developmental Biology, Institute for Plant Sciences and 12 Microbiology, University of Hamburg, 22609 Hamburg, Germany. 13 4 Present address : Nara Institute of Science and Technology, Graduate School of 14 Biological Sciences, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan 15 5 Institute for Integrative Biology of the Cell (I2BC), Commissariat à l’Energie 16 Atomique et aux Energies Alternatives (CEA), Centre National de la Recherche 17 Scientifique (CNRS), Université Paris-Sud, CEA-Saclay, Gif-sur-Yvette, France 18 * Corresponding author. [email protected] 19 1 bioRxiv preprint doi: https://doi.org/10.1101/606285; this version posted April 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Transcriptional Recapitulation and Subversion Of
Open Access Research2007KaiseretVolume al. 8, Issue 7, Article R131 Transcriptional recapitulation and subversion of embryonic colon comment development by mouse colon tumor models and human colon cancer Sergio Kaiser¤*, Young-Kyu Park¤†, Jeffrey L Franklin†, Richard B Halberg‡, Ming Yu§, Walter J Jessen*, Johannes Freudenberg*, Xiaodi Chen‡, Kevin Haigis¶, Anil G Jegga*, Sue Kong*, Bhuvaneswari Sakthivel*, Huan Xu*, Timothy Reichling¥, Mohammad Azhar#, Gregory P Boivin**, reviews Reade B Roberts§, Anika C Bissahoyo§, Fausto Gonzales††, Greg C Bloom††, Steven Eschrich††, Scott L Carter‡‡, Jeremy E Aronow*, John Kleimeyer*, Michael Kleimeyer*, Vivek Ramaswamy*, Stephen H Settle†, Braden Boone†, Shawn Levy†, Jonathan M Graff§§, Thomas Doetschman#, Joanna Groden¥, William F Dove‡, David W Threadgill§, Timothy J Yeatman††, reports Robert J Coffey Jr† and Bruce J Aronow* Addresses: *Biomedical Informatics, Cincinnati Children's Hospital Medical Center, Cincinnati, OH 45229, USA. †Departments of Medicine, and Cell and Developmental Biology, Vanderbilt University and Department of Veterans Affairs Medical Center, Nashville, TN 37232, USA. ‡McArdle Laboratory for Cancer Research, University of Wisconsin, Madison, WI 53706, USA. §Department of Genetics and Lineberger Cancer Center, University of North Carolina, Chapel Hill, NC 27599, USA. ¶Molecular Pathology Unit and Center for Cancer Research, Massachusetts deposited research General Hospital, Charlestown, MA 02129, USA. ¥Division of Human Cancer Genetics, The Ohio State University College of Medicine, Columbus, Ohio 43210-2207, USA. #Institute for Collaborative BioResearch, University of Arizona, Tucson, AZ 85721-0036, USA. **University of Cincinnati, Department of Pathology and Laboratory Medicine, Cincinnati, OH 45267, USA. ††H Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612, USA. ‡‡Children's Hospital Informatics Program at the Harvard-MIT Division of Health Sciences and Technology (CHIP@HST), Harvard Medical School, Boston, Massachusetts 02115, USA. -

BUB3 Antibody
Efficient Professional Protein and Antibody Platforms BUB3 Antibody Basic information: Catalog No.: UPA63666 Source: Rabbit Size: 50ul/100ul Clonality: Monoclonal Concentration: 1mg/ml Isotype: Rabbit IgG Purification: Protein A affinity purified Useful Information: WB:1:500-1:2000 ICC:1:50-1:200 Applications: IHC:1:50-1:200 IP:1:10-1:50 FC:1:50-1:100 Reactivity: Human, Mouse, Rat Specificity: This antibody recognizes BUB3 protein. Immunogen: Recombinant protein corresponding to the N-terminus of human Bub3. BUB3 (budding uninhibited by benzimidazoles 3 homolog), also known as BUB3L or hBUB3, is a conserved component of the mitotic spindle assembly complex (MCC). It contains five WD repeat domains and forms cell cycle constitutive complexes with BUB1 and BUBR1. BUB3 is essential for the ki- netochore localization of BUB1 and BUBR1. As a component of the MCC, BUB3 is involved in the essential spindle checkpoint pathway that operates Description: during early embryogenesis. The spindle checkpoint pathway functions to postpone the initiation of anaphase until chromosomes are properly at- tached to the spindle. This acts to ensure accurate chromosome segrega- tion. In addition, BUB3 plays a role in regulating the establishment of cor- rect kinetochore-microtubule attachments. BUB3 is also thought to bind Tctex1L (or DYNLT3), a dynein light chain. Uniprot: O43684(Human) Q9WVA3(Mouse) BiowMW: 37 kDa Buffer: 1*TBS (pH7.4), 1%BSA, 40%Glycerol. Preservative: 0.05% Sodium Azide. Storage: Store at 4°C short term and -20°C long term. Avoid freeze-thaw cycles. Note: For research use only, not for use in diagnostic procedure. Data: Gene Universal Technology Co. -
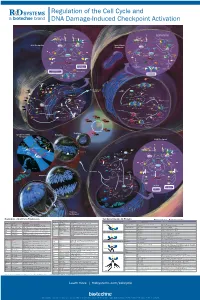
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

Control of the Cell Cycle During Meiotic Maturation of Mouse Oocytes
Control of the cell cycle during meiotic maturation of mouse oocytes Ibtissem Nabti Department of Cell and Developmental Biology University College London London, UK A thesis submitted for the degree of Doctor of Philosophy April 2010 Declaration The work presented in this thesis was conducted under the supervision of Professor John Carroll in the department of cell and developmental biology, University College London, and Professor Keith Jones in the department of reproductive physiology, University of Newcastle upon Tyne. I, Ibtissem Nabti confirm that the data presented in this thesis is my own and from original studies. Also, this dissertation has not previously been submitted, wholly or in part, to any other university for any degree or diploma. Ibtissem Nabti 1 Abstract The overall aim of the work presented in this thesis is to better understand the molecular mechanisms underlying the progression of the meiotic division in mammalian oocytes. The first goal of this project was to investigate the relative contribution of securin and cyclin B1 to the control of the protease separase during the indeterminate period of metaphase II arrest. I found here that although there are conditions in which either securin or MPF can prevent chromosome disjunction, such as during meiosis I, securin is the predominant inhibitor of separase during metaphase II arrest. Thus, CDK1 pharmacological inhibition as well as antibody inhibition of CDK1/cyclin B1 binding to separase, both failed to induce sister chromatid disjunction. Instead, securin morpholino knockdown induced sister chromatid separation, which could be rescued by injection of securin cRNA. I also examined the effect of CDK1 and MAPK activities on securin stability. -

Snapshot: the Nuclear Envelope II Andrea Rothballer and Ulrike Kutay Institute of Biochemistry, ETH Zurich, 8093 Zurich, Switzerland
SnapShot: The Nuclear Envelope II Andrea Rothballer and Ulrike Kutay Institute of Biochemistry, ETH Zurich, 8093 Zurich, Switzerland H. sapiens D. melanogaster C. elegans S. pombe S. cerevisiae Cytoplasmic filaments RanBP2 (Nup358) Nup358 CG11856 NPP-9 – – – Nup214 (CAN) DNup214 CG3820 NPP-14 Nup146 SPAC23D3.06c Nup159 Cytoplasmic ring and Nup88 Nup88 (Mbo) CG6819 – Nup82 SPBC13A2.02 Nup82 associated factors GLE1 GLE1 CG14749 – Gle1 SPBC31E1.05 Gle1 hCG1 (NUP2L1, NLP-1) tbd CG18789 – Amo1 SPBC15D4.10c Nup42 (Rip1) Nup98 Nup98 CG10198 Npp-10N Nup189N SPAC1486.05 Nup145N, Nup100, Nup116 Nup 98 complex RAE1 (GLE2) Rae1 CG9862 NPP-17 Rae1 SPBC16A3.05 Gle2 (Nup40) Nup160 Nup160 CG4738 NPP-6 Nup120 SPBC3B9.16c Nup120 Nup133 Nup133 CG6958 NPP-15 Nup132, Nup131 SPAC1805.04, Nup133 SPBP35G2.06c Nup107 Nup107 CG6743 NPP-5 Nup107 SPBC428.01c Nup84 Nup96 Nup96 CG10198 NPP-10C Nup189C SPAC1486.05 Nup145C Outer NPC scaffold Nup85 (PCNT1) Nup75 CG5733 NPP-2 Nup-85 SPBC17G9.04c Nup85 (Nup107-160 complex) Seh1 Nup44A CG8722 NPP-18 Seh1 SPAC15F9.02 Seh1 Sec13 Sec13 CG6773 Npp-20 Sec13 SPBC215.15 Sec13 Nup37 tbd CG11875 – tbd SPAC4F10.18 – Nup43 Nup43 CG7671 C09G9.2 – – – Centrin-21 tbd CG174931, CG318021 R08D7.51 Cdc311 SPCC1682.04 Cdc311 Nup205 tbd CG11943 NPP-3 Nup186 SPCC290.03c Nup192 Nup188 tbd CG8771 – Nup184 SPAP27G11.10c Nup188 Central NPC scaffold Nup155 Nup154 CG4579 NPP-8 tbd SPAC890.06 Nup170, Nup157 (Nup53-93 complex) Nup93 tbd CG7262 NPP-13 Nup97, Npp106 SPCC1620.11, Nic96 SPCC1739.14 Nup53(Nup35, MP44) tbd CG6540 NPP-19 Nup40 SPAC19E9.01c