PDF) If You Wish to Cite from It
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Genomes of Polyextremophilic Cyanidiales Contain 1% 2 Horizontally Transferred Genes with Diverse Adaptive Functions 3 4 Alessandro W
bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The genomes of polyextremophilic Cyanidiales contain 1% 2 horizontally transferred genes with diverse adaptive functions 3 4 Alessandro W. Rossoni1#, Dana C. Price2, Mark Seger3, Dagmar Lyska1, Peter Lammers3, 5 Debashish Bhattacharya4 & Andreas P.M. Weber1* 6 7 1Institute of Plant Biochemistry, Cluster of Excellence on Plant Sciences (CEPLAS), Heinrich 8 Heine University, Universitätsstraße 1, 40225 Düsseldorf, Germany 9 2Department of Plant Biology, Rutgers University, New Brunswick, NJ 08901, USA 10 3Arizona Center for Algae Technology and Innovation, Arizona State University, Mesa, AZ 11 85212, USA 12 4Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ 13 08901, USA 14 15 *Corresponding author: Prof. Dr. Andreas P.M. Weber, 16 e-mail: [email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 The role and extent of horizontal gene transfer (HGT) in eukaryotes are hotly disputed topics 20 that impact our understanding regarding the origin of metabolic processes and the role of 21 organelles in cellular evolution. -

Grapes for the Desert: Salt Stress Signaling in Vitis
Grapes for the Desert: Salt Stress Signaling in Vitis Zur Erlangung des akademischen Grades eines DOKTORS DER NATURWISSENSCHAFTEN (Dr. rer. Nat.) der Fakultät für Chemie und Biowissenschaften des Karlsruher Instituts für Technologie (KIT) genehmigte DISSERTATION von Ahmed Abd Alkarim Mohammed Ismail aus Damanhour, Ägypten Die vorliegende Dissertation wurde in der Abteilung Molekulare Zellbiologie, am Botanischen Institut des Karlsruher Instituts für Technologie (KIT) im Zeitraum von April 2010 bis Juli 2013 angefertigt. Dekan: Prof. Dr. M. Bastmeyer Referent: Prof. Dr. P. Nick Korreferent: Prof. Dr. H. Puchta Prof. Dr. Natalia Requena Dr. Christoph Basse Tag der mündlichen Prüfung: 19. Juli 2013 Hiermit erkläre ich, dass ich die vorliegende Dissertation, abgesehen von der Benutzung der angegebenen Hilfsmittel, selbstständig verfasst habe. Alle Stellen, die gemäß Wortlaut oder Inhalt aus anderen Arbeiten entnommen sind, wurden durch Angabe der Quelle als Entlehnungen kenntlich gemacht. Diese Dissertation liegt in gleicher oder ähnlicher Form keiner anderen Prüfungsbehörde vor. Karlsruhe, den 19. Juli 2013 Ahmed Abd Alkarim Mohammed Ismail Table of Contents Table of Contents Abbreviations ........................................................................... IV Summary / Zusammenfassung ............................................... XI 1 Introduction ............................................................................. 1 1.1 What does stress mean for a plant? .............................................. 1 1.2 Environmental -

Carnitine Metabolism to Trimethylamine by an Unusual Rieske-Type Oxygenase from Human Microbiota
Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota Yijun Zhua,1, Eleanor Jamesona,1, Marialuisa Crosattib,1, Hendrik Schäfera, Kumar Rajakumarb, Timothy D. H. Buggc, and Yin Chena,2 aSchool of Life Sciences and cDepartment of Chemistry, University of Warwick, Coventry CV4 7AL, United Kingdom; and bDepartment of Infection, Immunity, and Inflammation, University of Leicester, Leicester LE1 9HN, United Kingdom Edited by David W. Russell, University of Texas Southwestern Medical Center, Dallas, TX, and approved January 29, 2014 (received for review September 5, 2013) Dietary intake of L-carnitine can promote cardiovascular diseases in (14, 15). Assigning functions encoded in the human microbiome humans through microbial production of trimethylamine (TMA) using existing databases can be problematic. For example, the and its subsequent oxidation to trimethylamine N-oxide by hepatic Pfam protein database currently contains over 25% of protein flavin-containing monooxygenases. Although our microbiota are re- families with no assigned functions (release 26.0) (19). sponsible for TMA formation from carnitine, the underpinning mo- Lack of functional characterization of key microbial functions lecular and biochemical mechanisms remain unclear. In this study, in our microbiota is exemplified by very recent studies on car- using bioinformatics approaches, we first identified a two-component diovascular diseases (20–23). These studies have shown that the Rieske-type oxygenase/reductase (CntAB) and associated gene human microbiota is responsible for the production of trime- cluster proposed to be involved in carnitine metabolism in repre- thylamine N-oxide (TMAO), which is believed to promote ath- sentative genomes of the human microbiota. CntA belongs to a erogenesis through its interaction with macrophages and lipid group of previously uncharacterized Rieske-type proteins and has metabolism (20–23). -

Annual Conference Abstracts
ANNUAL CONFERENCE 14-17 April 2014 Arena and Convention Centre, Liverpool ABSTRACTS SGM ANNUAL CONFERENCE APRIL 2014 ABSTRACTS (LI00Mo1210) – SGM Prize Medal Lecture (LI00Tu1210) – Marjory Stephenson Climate Change, Oceans, and Infectious Disease Prize Lecture Dr. Rita R. Colwell Understanding the basis of antibiotic resistance University of Maryland, College Park, MD, USA as a platform for early drug discovery During the mid-1980s, satellite sensors were developed to monitor Laura JV Piddock land and oceans for purposes of understanding climate, weather, School of Immunity & Infection and Institute of Microbiology and and vegetation distribution and seasonal variations. Subsequently Infection, University of Birmingham, UK inter-relationships of the environment and infectious diseases Antibiotic resistant bacteria are one of the greatest threats to human were investigated, both qualitatively and quantitatively, with health. Resistance can be mediated by numerous mechanisms documentation of the seasonality of diseases, notably malaria including mutations conferring changes to the genes encoding the and cholera by epidemiologists. The new research revealed a very target proteins as well as RND efflux pumps, which confer innate close interaction of the environment and many other infectious multi-drug resistance (MDR) to bacteria. The production of efflux diseases. With satellite sensors, these relationships were pumps can be increased, usually due to mutations in regulatory quantified and comparatively analyzed. More recent studies of genes, and this confers resistance to antibiotics that are often used epidemic diseases have provided models, both retrospective and to treat infections by Gram negative bacteria. RND MDR efflux prospective, for understanding and predicting disease epidemics, systems not only confer antibiotic resistance, but altered expression notably vector borne diseases. -
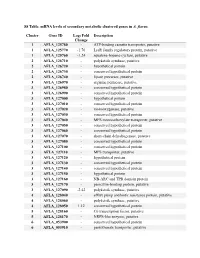
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Comparative Genomics of Bradyrhizobium Japonicum CPAC
Siqueira et al. BMC Genomics 2014, 15:420 http://www.biomedcentral.com/1471-2164/15/420 RESEARCH ARTICLE Open Access Comparative genomics of Bradyrhizobium japonicum CPAC 15 and Bradyrhizobium diazoefficiens CPAC 7: elite model strains for understanding symbiotic performance with soybean Arthur Fernandes Siqueira1,2†, Ernesto Ormeño-Orrillo3†,RangelCelsoSouza4,ElisetePainsRodrigues5, Luiz Gonzaga Paula Almeida4, Fernando Gomes Barcellos5, Jesiane Stefânia Silva Batista6, Andre Shigueyoshi Nakatani2, Esperanza Martínez-Romero3, Ana Tereza Ribeiro Vasconcelos4 and Mariangela Hungria1,2* Abstract Background: The soybean-Bradyrhizobium symbiosis can be highly efficient in fixing nitrogen, but few genomic sequences of elite inoculant strains are available. Here we contribute with information on the genomes of two commercial strains that are broadly applied to soybean crops in the tropics. B. japonicum CPAC 15 (=SEMIA 5079) is outstanding in its saprophytic capacity and competitiveness, whereas B. diazoefficiens CPAC 7 (=SEMIA 5080) is known for its high efficiency in fixing nitrogen. Both are well adapted to tropical soils. The genomes of CPAC 15 and CPAC 7 were compared to each other and also to those of B. japonicum USDA 6T and B. diazoefficiens USDA 110T. Results: Differences in genome size were found between species, with B. japonicum having larger genomes than B. diazoefficiens. Although most of the four genomes were syntenic, genome rearrangements within and between species were observed, including events in the symbiosis island. In addition to the symbiotic region, several genomic islands were identified. Altogether, these features must confer high genomic plasticity that might explain adaptation and differences in symbiotic performance. It was not possible to attribute known functions to half of the predicted genes. -

Thermophilic Bacteria Are Potential Sources of Novel Rieske Non-Heme
Chakraborty et al. AMB Expr (2017) 7:17 DOI 10.1186/s13568-016-0318-5 ORIGINAL ARTICLE Open Access Thermophilic bacteria are potential sources of novel Rieske non‑heme iron oxygenases Joydeep Chakraborty, Chiho Suzuki‑Minakuchi, Kazunori Okada and Hideaki Nojiri* Abstract Rieske non-heme iron oxygenases, which have a Rieske-type [2Fe–2S] cluster and a non-heme catalytic iron center, are an important family of oxidoreductases involved mainly in regio- and stereoselective transformation of a wide array of aromatic hydrocarbons. Though present in all domains of life, the most widely studied Rieske non-heme iron oxygenases are found in mesophilic bacteria. The present study explores the potential for isolating novel Rieske non- heme iron oxygenases from thermophilic sources. Browsing the entire bacterial genome database led to the identifi‑ cation of 45 homologs from thermophilic bacteria distributed mainly among Chloroflexi, Deinococcus–Thermus and Firmicutes. Thermostability, measured according to the aliphatic index, showed higher values for certain homologs compared with their mesophilic relatives. Prediction of substrate preferences indicated that a wide array of aromatic hydrocarbons could be transformed by most of the identified oxygenase homologs. Further identification of putative genes encoding components of a functional oxygenase system opens up the possibility of reconstituting functional thermophilic Rieske non-heme iron oxygenase systems with novel properties. Keywords: Rieske non-heme iron oxygenase, Oxidoreductase, Thermophiles, Aromatic hydrocarbons, Biotransformation Introduction of a wide array of agrochemically and pharmaceutically Rieske non-heme iron oxygenases (ROs) constitute a important compounds (Ensley et al. 1983; Wackett et al. large family of oxidoreductase enzymes involved primar- 1988; Hudlicky et al. -

2016 Joint Meeting Program
April 15 – 17, 2016 Fairmont Chicago Millennium Park • Chicago, Illinois The AAP/ASCI/APSA conference is jointly provided by Boston University School of Medicine and AAP/ASCI/APSA. Meeting Program and Abstracts www.jointmeeting.org www.jointmeeting.org Special Events at the 2016 AAP/ASCI/APSA Joint Meeting Friday, April 15 Saturday, April 16 ASCI President’s Reception ASCI Food and Science Evening 6:15 – 7:15 p.m. 6:30 – 9:00 p.m. Gold Room The Mid-America Club, Aon Center ASCI Dinner & New Member AAP Member Banquet Induction Ceremony (Ticketed guests only) (Ticketed guests only) 7:00 – 10:00 p.m. 7:30 – 9:45 p.m. Imperial Ballroom, Level B2 Rouge, Lobby Level How to Solve a Scientific Puzzle: Speaker: Clara D. Bloomfield, MD Clues from Stockholm and Broadway The Ohio State University Comprehensive Cancer Center Speaker: Joe Goldstein, MD APSA Welcome Reception & University of Texas Southwestern Medical Center at Dallas Presidential Address APSA Dinner (Ticketed guests only) 9:00 p.m. – Midnight Signature Room, 360 Chicago, 7:30 – 9:00 p.m. John Hancock Center (off-site) Rouge, Lobby Level Speaker: Daniel DelloStritto, APSA President Finding One’s Scientific Niche: Musings from a Clinical Neuroscientist Speaker: Helen Mayberg, MD, Emory University Dessert Reception (open to all attendees) 10:00 p.m. – Midnight Imperial Foyer, Level B2 Sunday, April 17 APSA Future of Medicine and www.jointmeeting.org Residency Luncheon Noon – 2:00 p.m. Rouge, Lobby Level 2 www.jointmeeting.org Program Contents General Program Information 4 Continuing Medical Education Information 5 Faculty and Speaker Disclosures 7 Scientific Program Schedule 9 Speaker Biographies 16 Call for Nominations: 2017 Harrington Prize for Innovation in Medicine 26 AAP/ASCI/APSA Joint Meeting Faculty 27 Award Recipients 29 Call for Nominations: 2017 Harrington Scholar-Innovator Award 31 Call for Nominations: George M. -

Purification and Characterization of a Novel Nitrilase of Rhodococcus
JOURNAL OF BACTERIOLOGY, Sept. 1990, p. 4807-4815 Vol. 172, No. 9 0021-9193/90/094807-09$02.00/0 Copyright X3 1990, American Society for Microbiology Purification and Characterization of a Novel Nitrilase of Rhodococcus rhodochrous K22 That Acts on Aliphatic Nitriles MICHIHIKO KOBAYASHI,* NORIYUKI YANAKA, TORU NAGASAWA, AND HIDEAKI YAMADA Department ofAgricultural Chemistry, Faculty ofAgriculture, Kyoto University, Sakyo-ku, Kyoto 606, Japan Received 18 April 1990/Accepted 8 June 1990 A novel nitrilase that preferentially catalyzes the hydrolysis of aliphatic nitriles to the corresponding carboxylic acids and ammonia was found in the cells of a facultative crotononitrile-utilizing actinomycete isolated from soil. The strain was taxonomically studied and identified as Rhodococcus rhodochrous. The nitrilase was purified, with 9.08% overall recovery, through five steps from a cell extract of the stain. After the last step, the purified enzyme appeared to be homogeneous, as judged by polyacrylamide gel electrophoresis, analytical centrifugation, and double immunodiffusion in agarose. The relative molecular weight values for the native enzyme, estimated from the ultracentrifugal equilibrium and by high-performance liquid chromatog- raphy, were approximately 604,000 + 30,000 and 650,000, respectively, and the enzyme consisted of 15 to 16 subunits identical in molecular weight (41,000). The enzyme acted on aliphatic olefinic nitriles such as crotononitrile and acrylonitrile as the most suitable substrates. The apparent Km values for crotononitrile and acrylonitrile were 18.9 and 1.14 mM, respectively. The nitrilase also catalyzed the direct hydrolysis of saturated aliphatic nitriles, such as valeronitrile, 4-chlorobutyronitrile, and glutaronitrile, to the correspond- ing acids without the formation of amide intermediates. -

Direct Link to Fulltext
ISOLATION, IDENTIFICATION AND SUBSTRATE SPECIFICITY OF A NITRILASE PRODUCING BACTERIA, Acidovorax sp. SK1 Soumya Koippully, Subha Swaraj Pattnaik, Siddhardha Busi* Address(es): Department of Microbiology, School of Life Sciences, Pondicherry University, Puducherry-605 014, India. *Corresponding author: [email protected] doi: 10.15414/jmbfs.2018.8.2.788-793 ARTICLE INFO ABSTRACT Received 13. 5. 2018 The process of biocatalysis or biotransformation remains the core of industrial biotechnology owing to its importance in the synthesis of Revised 3. 9. 2018 high-value products in a cost effective manner. Nitrile biotransformation has also achieved considerable attention in last few decades Accepted 5. 9. 2018 due to its widespread biotechnological and industrial applications. In the present study, a versatile, potent nitrile-degrading bacterium, Published 1. 10. 2018 Acidovorax sp. SK1 was isolated from the cracker waste dumping site of Sivakasi, Tamilnadu and characterized for its biocatalytic potential, nitrilase production and enzyme activity. A pH sensitive indicator-based assay was performed to identify the nitrile degrading ability of the isolated samples. Semi quantitative High performance thin layer chromatographic (HPTLC) method was performed for Regular article quantitative measurement of mandelic acid produced from degradation of nitrile compound, mandelonitrile. The optimization of medium and nutritional parameters were studied for the improvement of nitrilase activity, which indicated that maximum nitrilase activity was observed at an optimum pH of 7.0, agitation at 100 rpm, glucose as best carbon source (10 g/L) and yeast extract (0.1 g/L) as principal nitrogen source. Biomass is also a critical parameter in the biocatalysis of mandelonitrile to mandelic acid and at a biomass of 100 mg/L, maximum nitrilase activity of 0.026 I.U was observed. -

Role in Plant Stress Physiology and Regulation of Gene Expression
Characterisation of selected Arabidopsis aldehyde dehydrogenase genes: role in plant stress physiology and regulation of gene expression Dissertation Zur Erlangung des Doktorgrades (Dr. rer. nat.) der Mathematisch-Naturwissenschaftlichen Fakultät der Rheinischen Friedrich-Wilhelms-Universität Bonn vorgelegt von Tagnon Dègbédji MISSIHOUN aus Cotonou, Benin Bonn, November 2010 Angefertigt mit Genehmigung der Mathematisch- Naturwissenschaftlichen Fakultät der Rheinischen Friedrich-Wilhelms-Universität Bonn Gedruckt mit Unterstützung des Deutschen Akademischen Austauschdienstes 1. Referentin: Prof. Dr. Dorothea Bartels 2. Koreferent: Priv. Doz. Dr. Hans-Hubert Kirch Tag der Promotion: 22. Februar 2011 Erscheinungsjahr: 2011 II DECLARATION I hereby declare that the whole PhD thesis is my own work, except where explicitly stated otherwise in the text or in the bibliography. Bonn, November 2010 ------------------------------------ Tagnon D. MISSIHOUN III DEDICATION To My wife: Fabienne TOSSOU-MISSIHOUN and our kids Floriane S. Jennifer and Sègnon Anges- Anis My parents: Lucrèce KOTOMALE and Dadjo MISSIHOUN My sister and brothers: Mariette, Marius, Ricardo, Renaud, Ulrich And my dearest aunts and uncles: Hoho, Rebecca, Cyriaque, Dominique, Alphonsine IV CONTENTS ABBREVIATIONS ...............................................................................................................................................X FIGURES AND TABLES ...............................................................................................................................XIII -

Sample Thesis Title with a Concise and Accurate
DISTINCT FUNCTIONS FOR THE HIGHLY RELATED PP2A B55 REGULATORY SUBUNITS (Bα AND Bδ) IN CELL CYCLE REGULATION AND TUMOURIGENESIS by Dominik Sommerfeld B.Sc., The University of British Columbia, 2009 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE AND POSTDOCTORAL STUDIES (Experimental Medicine) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) August 2018 © Dominik Sommerfeld, 2018 The following individuals certify that they have read, and recommend to the Faculty of Graduate and Postdoctoral Studies for acceptance, the dissertation entitled: Distinct functions for the highly related PP2A B55 regulatory subunits (Bα and Bδ) in cell cycle regulation and tumourigenesis submitted by Dominik Sommerfeld in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Experimental Medicine Examining Committee: Catherine Pallen, Ph.D. Supervisor Christopher Maxwell, Ph.D. Supervisory Committee Member Gregg Morin, Ph.D. Supervisory Committee Member Stephan Taubert, Ph.D. University Examiner Calvin Roskelley, Ph.D. University Examiner Additional Supervisory Committee Members: Samuel Aparicio, Ph.D. Supervisory Committee Member ii Abstract Progression through the phases of the cell cycle (G1, S, G2, and mitosis) is largely driven by the coordinated activities of protein kinases and phosphatases, which orchestrate the phase-specific phosphorylation of proteins. Protein Phosphatase 2A (PP2A) – a heterotrimeric holoenzyme complex composed of a scaffold (A), catalytic (C), and variable regulatory (B) subunit – has emerged as an essential cell cycle regulator. Specifically, PP2A complexes containing the B55 family of regulatory subunits (PP2A-B55) have been implicated in the control of various cell cycle phases. My studies investigated isoform-specific roles of the two highly related and abundantly expressed B55 subunits, Bα and Bδ, in the regulation of G1, S, G2 phase and mitotic progression.