Toxicogenomics of Kojic Acid on Gene Expression Profiling of A375
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Order Estimation Based Approach to Identify Response Genes
AN ORDER ESTIMATION BASED APPROACH TO IDENTIFY RESPONSE GENES FOR MICRO ARRAY TIME COURSE DATA A Thesis Presented to The Faculty of Graduate Studies of The University of Guelph by ZHIHENG LU In partial fulfilment of requirements for the degree of Doctor of Philosophy September, 2008 © Zhiheng Lu, 2008 Library and Bibliotheque et 1*1 Archives Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A0N4 Ottawa ON K1A0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-47605-5 Our file Notre reference ISBN: 978-0-494-47605-5 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library permettant a la Bibliotheque et Archives and Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par Plntemet, prefer, telecommunication or on the Internet, distribuer et vendre des theses partout dans loan, distribute and sell theses le monde, a des fins commerciales ou autres, worldwide, for commercial or non sur support microforme, papier, electronique commercial purposes, in microform, et/ou autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in et des droits moraux qui protege cette these. this thesis. Neither the thesis Ni la these ni des extraits substantiels de nor substantial extracts from it celle-ci ne doivent etre imprimes ou autrement may be printed or otherwise reproduits sans son autorisation. -

Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-Like Mouse Models: Tracking the Role of the Hairless Gene
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 5-2006 Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene Yutao Liu University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Life Sciences Commons Recommended Citation Liu, Yutao, "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino- like Mouse Models: Tracking the Role of the Hairless Gene. " PhD diss., University of Tennessee, 2006. https://trace.tennessee.edu/utk_graddiss/1824 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Yutao Liu entitled "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the requirements for the degree of Doctor of Philosophy, with a major in Life Sciences. Brynn H. Voy, Major Professor We have read this dissertation and recommend its acceptance: Naima Moustaid-Moussa, Yisong Wang, Rogert Hettich Accepted for the Council: Carolyn R. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

| Hai Lala at Matalamitaka Huoleht I
|HAI LALA AT MATALAMITAKAUS009816096B2 HUOLEHT I (12 ) United States Patent (10 ) Patent No. : US 9 ,816 , 096 B2 Heintz et al. (45 ) Date of Patent: Nov . 14 , 2017 ( 54 ) METHODS AND COMPOSITIONS FOR 6 , 143 , 566 A 11/ 2000 Heintz et al. TRANSLATIONAL PROFILING AND 6 , 156 , 574 A 12 / 2000 Heintz et al. 6 , 252 , 130 B1 6 / 2001 Federoff MOLECULAR PHENOTYPING 6 , 270, 969 B1 8 / 2001 Hartley et al. 6 , 403 ,374 B1 6 / 2002 Tsien et al. (71 ) Applicant: THE ROCKEFELLER 6 , 410 , 317 B1 6 /2002 Farmer UNIVERSITY , New York , NY (US ) 6 , 441 , 269 B1 8 / 2002 Serafini et al . 6 , 485 , 912 B1 11/ 2002 Heintz et al. @ 6 , 495 , 318 B2 12 / 2002 Harney ( 72 ) Inventors: Nathaniel Heintz , Pelham Manor, NY 6 ,635 ,422 B2 10 / 2003 Keene et al. (US ) ; Paul Greengard , New York , NY 6 , 821, 759 B1 11/ 2004 Heintz et al . (US ) ; Myriam Heiman , New York , NY 7 , 098, 031 B2B2 8 /2006 Choulika et al . (US ) ; Anne Schaefer , New York , NY 7 ,297 ,482 B2 11 /2007 Anderson et al . (US ) ; Joseph P . Doyle , New York , NY 7 , 393 , 632 B2 7 / 2008 Cheo et al. 2003 /0119104 A1 6 /2003 Perkins et al . (US ) ; Joseph D . Dougherty , St. Louis , 2004 / 0023256 A1 2 / 2004 Puglisi et al . MO (US ) 2005 / 0009028 Al 1 /2005 Heintz et al. 2006 /0183147 AL 8 /2006 Meyer - Franke (73 ) Assignee : THE ROCKEFELLER 2011/ 0314565 Al 12 /2011 Heintz et al . UNIVERSITY , New York , NY (US ) FOREIGN PATENT DOCUMENTS ( * ) Notice : Subject to any disclaimer , the term of this patent is extended or adjusted under 35 EP 1132479 A1 9 / 2001 WO WO -01 / 48480 A1 7 /2001 U . -

Estrogen Receptor Α Polymorphism in a Species with Alternative Behavioral Phenotypes
Estrogen receptor α polymorphism in a species with alternative behavioral phenotypes Brent M. Hortona, William H. Hudsonb, Eric A. Ortlundb, Sandra Shirka, James W. Thomasc, Emily R. Youngd, Wendy M. Zinzow-Kramera, and Donna L. Maneya,1 aDepartment of Psychology, Emory University, Atlanta, GA 30322; bDepartment of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322; cNIH Intramural Sequencing Center, National Human Genome Research Institute, National Institutes of Health, Rockville, MD 20852; and dDepartment of Biology, Georgia Institute of Technology, Atlanta, GA 30332 Edited by Ellen D. Ketterson, Indiana University, Bloomington, IN, and accepted by the Editorial Board December 5, 2013 (received for review September 12, 2013) The evolution of behavior relies on changes at the level of the (7, 8). Morph differences in behavior cannot be entirely explained genome; yet the ability to attribute a behavioral change to by these hormones, however, because the differences persist even a specific, naturally occurring genetic change is rare in vertebrates. when plasma levels are experimentally equalized (9). Individual In the white-throated sparrow (Zonotrichia albicollis), a chromo- variation in steroid-dependent behavior may be better explained somal polymorphism (ZAL2/2m) is known to segregate with a be- by neural sensitivity to the hormones, for example by variation in havioral phenotype. Individuals with the ZAL2m haplotype engage the distribution and abundance of steroid receptors (10, 11). in more territorial aggression and less parental behavior than indi- The gene encoding estrogen receptor α (ERα), estrogen re- m viduals without it. These behaviors are thought to be mediated by ceptor 1 (ESR1), has been captured by the ZAL2 rearrange- sensitivity to sex steroids, and the chromosomal rearrangement ment (3) and has therefore likely differentiated between the underlying the polymorphism has captured a prime candidate haplotypes. -

Valproic Acid Reversed Pathologic Endothelial Cell Gene Expression Profile Associated with Ischemia– Reperfusion Injury in a Swine Hemorrhagic Shock Model
From the Peripheral Vascular Surgery Society Valproic acid reversed pathologic endothelial cell gene expression profile associated with ischemia– reperfusion injury in a swine hemorrhagic shock model Marlin Wayne Causey, MD,a Shashikumar Salgar, PhD,b Niten Singh, MD,a Matthew Martin, MD,a and Jonathan D. Stallings, PhD,b Tacoma, Wash Background: Vascular endothelial cells serve as the first line of defense for end organs after ischemia and reperfusion injuries. The full etiology of this dysfunction is poorly understood, and valproic acid (VPA) has proven to be beneficial after traumatic injury. The purpose of this study was to determine the mechanism of action through which VPA exerts its beneficial effects. Methods: Sixteen Yorkshire swine underwent a standardized protocol for an ischemia–reperfusion injury through received (6 ؍ hemorrhage and a supraceliac cross-clamp with ensuing 6-hour resuscitation. The experimental swine (n Aortic .(5 ؍ and injury-control models (n (5 ؍ VPA at cross-clamp application and were compared with sham (n endothelium was harvested, and microarray analysis was performed along with a functional clustering analysis with gene transcript validation using relative quantitative polymerase chain reaction. Results: Clinical comparison of experimental swine matched for sex, weight, and length demonstrated that VPA significantly decreased resuscitative requirements, with improved hemodynamics and physiologic laboratory measure- ments. Six transcript profiles from the VPA treatment were compared with the 1536 gene transcripts (529 up and 1007 down) from sham and injury-control swine. Microarray analysis and a Database for Annotation, Visualization and Integrated Discovery functional pathway analysis approach identified biologic processes associated with pathologic vascular endothelial function, specifically through functional cluster pathways involving apoptosis/cell death and angiogenesis/vascular development, with five specific genes (THBS1, TNFRSF12A, ANGPTL4, RHOB, and RTN4) identified as members of both functional clusters. -

Pirfenidone Is Renoprotective in Diabetic Kidney Disease
BASIC RESEARCH www.jasn.org Pirfenidone Is Renoprotective in Diabetic Kidney Disease ʈ Satish P. RamachandraRao,*†‡ Yanqing Zhu,‡ Timothy Ravasi,§ Tracy A. McGowan,‡ Irene Toh,‡ Stephen R. Dunn,‡¶ Shinichi Okada,*† Michael A. Shaw,** and Kumar Sharma*†‡ *Center for Renal Translational Medicine, Division of Nephrology-Hypertension, Department of Medicine, and ʈ §Department of Bioengineering, Jacobs School of Engineering, University of California, San Diego, Scripps NeuroAIDS Preclinical Studies Centre, and †Veterans Administration San Diego Healthcare System, La Jolla, California, ‡Center for Novel Therapies in Kidney Disease, Department of Medicine, ¶Cancer Genomics Facility, Kimmel Cancer Center, and **Proteomics and Mass Spectrometry Core Facility, Department of Cancer Biology, Thomas Jefferson University, Philadelphia, Pennsylvania ABSTRACT Although several interventions slow the progression of diabetic nephropathy, current therapies do not halt progression completely. Recent preclinical studies suggested that pirfenidone (PFD) prevents fibrosis in various diseases, but the mechanisms underlying its antifibrotic action are incompletely understood. Here, we evaluated the role of PFD in regulation of the extracellular matrix. In mouse mesangial cells, PFD decreased TGF- promoter activity, reduced TGF- protein secretion, and inhibited TGF-–induced Smad2-phosphor- ylation, 3TP-lux promoter activity, and generation of reactive oxygen species. To explore the therapeutic potential of PFD, we administered PFD to 17-wk-old db/db mice for 4 wk. PFD treatment significantly reduced mesangial matrix expansion and expression of renal matrix genes but did not affect albuminuria. Using liquid chromatography with subsequent electrospray ionization tandem mass spectrometry, we iden- tified 21 proteins unique to PFD-treated diabetic kidneys. Analysis of gene ontology and protein–protein interactions of these proteins suggested that PFD may regulate RNA processing. -

Prediction of Tissue-Specific Cis-Regulatory Sequences: Application to the Ascidian Ciona Intestinalis and the Anterior Neurectoderm Maximilian Häussler
Prediction of tissue-specific cis-regulatory sequences: application to the ascidian Ciona intestinalis and the anterior neurectoderm Maximilian Häussler To cite this version: Maximilian Häussler. Prediction of tissue-specific cis-regulatory sequences: application to the ascidian Ciona intestinalis and the anterior neurectoderm. Cellular Biology. Université Paris Sud - Paris XI, 2009. English. tel-00413501 HAL Id: tel-00413501 https://tel.archives-ouvertes.fr/tel-00413501 Submitted on 4 Sep 2009 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Université Paris XI Discipline Biologie Cellulaire et Moléculaire École doctorale Gènes, Génomes, Cellules Thèse pour obtenir le grade de Docteur de l'Université Paris XI Soutenance prévu le 15. Juillet 2009 par Maximilian Häussler Prédiction des séquences cis-regulatrices tissu-spéci- fiques: application à l'ascidie Ciona intestinalis et au neurectoderme antérieur Prediction of tissue-specific cis-regulatory sequences: application to the ascidian Ciona intestinalis and the anterior neurectoderm Jury President M. Pierre Capy Rapporteurs: M. Nicolas Pollet M. Sebastian Shimeld Examinateur: M. Elia Stupka Directeur de thèse: M. Jean-Stéphane Joly This thesis can be downloaded from http://hal.archives-ouvertes.fr as a PDF file Summary The detection and annotation of cis-regulatory sequences is a difficult problem. -

Evolutionary Fate of Retroposed Gene Copies in the Human Genome
Evolutionary fate of retroposed gene copies in the human genome Nicolas Vinckenbosch*, Isabelle Dupanloup*†, and Henrik Kaessmann*‡ *Center for Integrative Genomics, University of Lausanne, Ge´nopode, 1015 Lausanne, Switzerland; and †Computational and Molecular Population Genetics Laboratory, Zoological Institute, University of Bern, 3012 Bern, Switzerland Communicated by Wen-Hsiung Li, University of Chicago, Chicago, IL, December 30, 2005 (received for review December 14, 2005) Given that retroposed copies of genes are presumed to lack the and rodent genomes (7–12). In addition, three recent studies regulatory elements required for their expression, retroposition using EST data (13, 14) and tiling-microarray data from chro- has long been considered a mechanism without functional rele- mosome 22 (15) indicated that retrocopy transcription may be vance. However, through an in silico assay for transcriptional widespread, although these surveys were limited, and potential activity, we identify here >1,000 transcribed retrocopies in the functional implications were not addressed. human genome, of which at least Ϸ120 have evolved into bona To further explore the functional significance of retroposition fide genes. Among these, Ϸ50 retrogenes have evolved functions in the human genome, we systematically screened for signatures in testes, more than half of which were recruited as functional of selection related to retrocopy transcription. Our results autosomal counterparts of X-linked genes during spermatogene- suggest that retrocopy transcription is not rare and that the sis. Generally, retrogenes emerge ‘‘out of the testis,’’ because they pattern of transcription of human retrocopies has been pro- are often initially transcribed in testis and later evolve stronger and foundly shaped by natural selection, acting both for and against sometimes more diverse spatial expression patterns. -

Identification of Bleomycin and Radiation-Induced Pulmonary Fibrosis Susceptibility Genes in Mice Anne-Marie Lemay Department Of
Identification of bleomycin and radiation-induced pulmonary fibrosis susceptibility genes in mice Anne-Marie Lemay Department of Human Genetics McGill University, Montréal February 4th, 2010 A thesis submitted to McGill University in partial fulfilment of the requirements of the degree of Doctor of Philosophy © Anne-Marie Lemay 2010 Comme il est profond, ce mystère de l’Invisible ! Nous ne pouvons le sonder avec nos sens misérables, avec nos yeux qui ne savent apercevoir ni le trop petit, ni le trop grand, ni le trop près, ni le trop loin, ni les habitants d’une étoile, ni les habitants d’une goutte d’eau… Guy de Maupassant Le Horla ii Table of contents Table of contents ................................................................................................... iii Abstract...................................................................................................................vi Résumé ................................................................................................................ viii Acknowledgments................................................................................................... x Abbreviations........................................................................................................ xii Original contributions to knowledge...................................................................xiv Author contribution to research...........................................................................xv List of figures ........................................................................................................ -
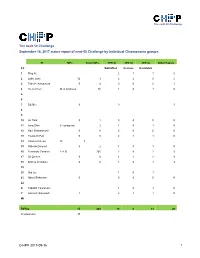
Next-MP50 Status Report
The neXt-50 Challenge The neXt-50 Challenge September 16, 2017 status report of next-50 Challenge by individual Chromosome groups. PI MPs Silver MPs JPR SI JPR SI JPR SI Other Papers Ch Submitted In press In revision 1 Ping Xu 2 1 1 0 2 Lydie Lane 12 3 2 2 0 2 3 Takeshi Kawamura 0 4 0 0 0 1 4 Yu-Ju Chen 26 in progress 86 1 0 1 0 5 6 7 Ed Nice 0 0 1 8 9 10 Jin Park 0 1 0 0 0 0 11 Jong Shin 2 in progress 2 1 0 1 0 12 Ravi Sirdeshmukh 0 0 0 0 0 0 13 Young-Ki Paik 0 5 2 1 1 0 14 CharLes Pineau 12 3 15 GiLberto Domont 0 2 1 0 1 0 16 Fernando CorraLes 2 (+ 9) 165 2 0 2 5 17 GiL Omenn 0 0 3 1 2 8 18 Andrey Archakov 0 0 1 0 1 3 19 20 Siqi Liu 1 0 1 21 Albert Sickmann 0 0 0 0 0 22 X Tadashi Yamamoto 1 0 1 0 Y Hosseini SaLekdeh 1 2 1 1 0 Mt TOTAL 15 268 19 6 13 20 (in progress) 37 C-HPP 2017-09-16 1 The neXt-50 Challenge Chromosome 1 (Ping Xu) PIC Leaders: Ping Xu, Fuchu He Contributing labs: Ping Xu, Beijing Proteome Research Center Fuchu He, Beijing Proteome Research Center Dong Yang, Beijing Proteome Research Center Wantao Ying, Beijing Proteome Research Center Pengyuan Yang, Fudan University Siqi Liu, Beijing Genome Institute Qinyu He, Jinan University Major lab members or partners contributing to the neXt50: Yao Zhang (Beijing Proteome Research Center), Yihao Wang (Beijing Proteome Research Center), Cuitong He (Beijing Proteome Research Center), Wei Wei (Beijing Proteome Research Center), Yanchang Li (Beijing Proteome Research Center), Feng Xu (Beijing Proteome Research Center), Xuehui Peng (Beijing Proteome Research Center).