Increased Expression of Neutrophil-Related Genes in Patients with Early Sepsis-Induced ARDS
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
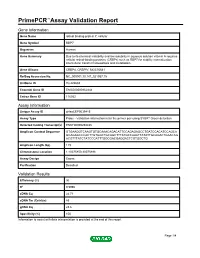
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name retinol binding protein 7, cellular Gene Symbol RBP7 Organism Human Gene Summary Due to its chemical instability and low solubility in aqueous solution vitamin A requires cellular retinol-binding proteins (CRBPs) such as RBP7 for stability internalization intercellular transfer homeostasis and metabolism. Gene Aliases CRBP4, CRBPIV, MGC70641 RefSeq Accession No. NC_000001.10, NT_021937.19 UniGene ID Hs.422688 Ensembl Gene ID ENSG00000162444 Entrez Gene ID 116362 Assay Information Unique Assay ID qHsaCEP0039415 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000294435 Amplicon Context Sequence GTGAAGGTCAAGTGTGCAAACAGACATTCCAGAGAGCCTGATCCACATCCAGCA GCAGAGCCCACTTGTGGCTGCAGCTTTATGCCAAATTATATTGCAGACTGAACAG ACGTTTATCTATCCCATTTGGCGACGAGGACTCGTGGCTG Amplicon Length (bp) 119 Chromosome Location 1:10075850-10075998 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 96 R2 0.9996 cDNA Cq 24.71 cDNA Tm (Celsius) 83 gDNA Cq 23.6 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report RBP7, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Vertebrate Fatty Acid and Retinoid Binding Protein Genes and Proteins: Evidence for Ancient and Recent Gene Duplication Events
In: Advances in Genetics Research. Volume 11 ISBN: 978-1-62948-744-1 Editor: Kevin V. Urbano © 2014 Nova Science Publishers, Inc. Chapter 7 Vertebrate Fatty Acid and Retinoid Binding Protein Genes and Proteins: Evidence for Ancient and Recent Gene Duplication Events Roger S. Holmes Eskitis Institute for Drug Discovery and School of Biomolecular and Physical Sciences, Griffith University, Nathan, QLD, Australia Abstract Fatty acid binding proteins (FABP) and retinoid binding proteins (RBP) are members of a family of small, highly conserved cytoplasmic proteins that function in binding and facilitating the cellular uptake of fatty acids, retinoids and other hydrophobic compounds. Several human FABP-like genes are expressed in the body: FABP1 (liver); FABP2 (intestine); FABP3 (heart and skeletal muscle); FABP4 (adipocyte); FABP5 (epidermis); FABP6 (ileum); FABP7 (brain); FABP8 (nervous system); FABP9 (testis); and FABP12 (retina and testis). A related gene (FABP10) is expressed in lower vertebrate liver and other tissues. Four RBP genes are expressed in human tissues: RBP1 (many tissues); RBP2 (small intestine epithelium); RBP5 (kidney and liver); and RBP7 (kidney and heart). Comparative FABP and RBP amino acid sequences and structures and gene locations were examined using data from several vertebrate genome projects. Sequence alignments, key amino acid residues and conserved predicted secondary and tertiary structures were also studied, including lipid binding regions. Vertebrate FABP- and RBP- like genes usually contained 4 coding exons in conserved locations, supporting a common evolutionary origin for these genes. Phylogenetic analyses examined the relationships and evolutionary origins of these genes, suggesting division into three FABP gene classes: 1: FABP1, FABP6 and FABP10; 2: FABP2; and 3, with 2 groups: 3A: FABP4, FABP8, FABP9 and FABP12; and 3B: and FABP3, FABP5 and FABP7. -

Cancer Informatics 2014:13 13–20 Doi: 10.4137/CIN.S13495
Open Access: Full open access to Cancer this and thousands of other papers at http://www.la-press.com. Informatics OmicCircos: A Simple-to-Use R Package for the Circular Visualization of Multidimensional Omics Data Ying Hu, Chunhua Yan, Chih-Hao Hsu, Qing-Rong Chen, Kelvin Niu, George A. Komatsoulis and Daoud Meerzaman Center for Biomedical Informatics and Information Technology, National Cancer Institute, Rockville, MD 20850, USA. ABSTRACT SUMMarY: OmicCircos is an R software package used to generate high-quality circular plots for visualizing genomic variations, including mutation patterns, copy number variations (CNVs), expression patterns, and methylation patterns. Such variations can be displayed as scatterplot, line, or text-label figures. Relationships among genomic features in different chromosome positions can be represented in the forms of polygons or curves. Utilizing the statistical and graphic functions in an R/Bioconductor environment, OmicCircos performs statistical analyses and displays results using cluster, boxplot, histogram, and heatmap formats. In addition, OmicCircos offers a number of unique capabilities, including independent track drawing for easy modifica- tion and integration, zoom functions, link-polygons, and position-independent heatmaps supporting detailed visualization. AVaiLabiLitY AND IMPLEMENtatiON: OmicCircos is available through Bioconductor at http://www.bioconductor.org/packages/devel/bioc/ html/OmicCircos.html. An extensive vignette in the package describes installation, data formatting, and workflow procedures. The software is open source under the Artistic−2.0 license. KEYWORDS: R package, circular plot, genomic variation CITATION: Hu et al. OmicCircos: A Simple-to-Use R Package for the Circular Visualization of Multidimensional Omics Data. Cancer Informatics 2014:13 13–20 doi: 10.4137/CIN.S13495. -

(LCA9) for Leber's Congenital Amaurosis on Chromosome 1P36
European Journal of Human Genetics (2003) 11, 420–423 & 2003 Nature Publishing Group All rights reserved 1018-4813/03 $25.00 www.nature.com/ejhg SHORT REPORT Identification of a locus (LCA9) for Leber’s congenital amaurosis on chromosome 1p36 T Jeffrey Keen1, Moin D Mohamed1, Martin McKibbin2, Yasmin Rashid3, Hussain Jafri3, Irene H Maumenee4 and Chris F Inglehearn*,1 1Molecular Medicine Unit, University of Leeds, Leeds, UK; 2Department of Ophthalmology, St James’s University Hospital, Leeds, UK; 3Department of Obstetrics and Gynaecology, Fatima Jinnah Medical College, Lahore, Pakistan; 4Department of Ophthalmology, Johns Hopkins University School of Medicine, Baltimore, MD, USA Leber’s congenital amaurosis (LCA) is the most common cause of inherited childhood blindness and is characterised by severe retinal degeneration at or shortly after birth. We have identified a new locus, LCA9, on chromosome 1p36, at which the disease segregates in a single consanguineous Pakistani family. Following a whole genome linkage search, an autozygous region of 10 cM was identified between the markers D1S1612 and D1S228. Multipoint linkage analysis generated a lod score of 4.4, strongly supporting linkage to this region. The critical disease interval contains at least 5.7 Mb of DNA and around 50 distinct genes. One of these, retinoid binding protein 7 (RBP7), was screened for mutations in the family, but none was found. European Journal of Human Genetics (2003) 11, 420–423. doi:10.1038/sj.ejhg.5200981 Keywords: Leber’s congenital amaurosis; LCA; LCA9; retina; linkage; 1p36 Introduction guineous pedigrees, known as homozygosity or autozygos- Leber’s congenital amaurosis (LCA) is the name given to a ity mapping, is a powerful approach to identify recessively group of recessively inherited retinal dystrophies repre- inherited disease gene loci.10 Cultural precedents in some senting the most common genetic cause of blindness in Pakistani communities have led to a high frequency of infants and children. -

NIH Public Access Author Manuscript Arterioscler Thromb Vasc Biol
NIH Public Access Author Manuscript Arterioscler Thromb Vasc Biol. Author manuscript; available in PMC 2011 March 1. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Arterioscler Thromb Vasc Biol. 2010 March ; 30(3): 518±525. doi:10.1161/ATVBAHA.109.200733. Bioinformatic Analysis of Gene Sets Regulated by Ligand- Activated and Dominant-Negative PPARγ in Mouse Aorta Henry L. Keen1, Carmen M. Halabi2, Andreas M. Beyer2, Willem J. de Lange1, Xuebo Liu1, Nobuyo Maeda9, Frank M. Faraci1,5, Thomas L. Casavant6,7,8, and Curt D. Sigmund1,3,4 1Department of Internal Medicine, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA, USA 2Genetics Graduate Program, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA, USA 3Department of Molecular Physiology and Biophysics, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA, USA 4Center on Functional Genomics of Hypertension, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA, USA 5Department of Pharmacology, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA, USA 6Center for Bioinformatics and Computational Biology, University of Iowa, Iowa City, IA, USA 7 Department of Electrical and Computer Engineering, University of Iowa, Iowa City, IA, USA 8 Department of Biomedical Engineering, University of Iowa, Iowa City, IA, USA 9Department of Pathology, University of North Carolina, Chapel Hill, NC, USA Abstract Objective—Drugs that activate PPARγ improve glucose sensitivity and lower blood pressure, whereas dominant negative mutations in PPARγ cause severe insulin resistance and hypertension. -

The Identification of Novel Regions for Reproduction Trait in Landrace and Large White Pigs Using a Single Step Genome-Wide Association Study
Open Access Asian-Australas J Anim Sci Vol. 31, No. 12:1852-1862 December 2018 https://doi.org/10.5713/ajas.18.0072 pISSN 1011-2367 eISSN 1976-5517 The identification of novel regions for reproduction trait in Landrace and Large White pigs using a single step genome-wide association study Rattikan Suwannasing1, Monchai Duangjinda1,*, Wuttigrai Boonkum1, Rutjawate Taharnklaew2, and Komson Tuangsithtanon3 * Corresponding Author: Monchai Duangjinda Objective: The purpose of this study was to investigate a single step genome-wide association Tel: +66-43-202362, Fax: +66-43-202361, E-mail: [email protected] study (ssGWAS) for identifying genomic regions affecting reproductive traits in Landrace and Large White pigs. 1 Department of Animal Science, Faculty of Agriculture, Methods: The traits included the number of pigs weaned per sow per year (PWSY), the Khon Kaen University, Khon Kaen 40002, Thailand 2 Research and Development Center Betagro Group, number of litters per sow per year (LSY), pigs weaned per litters (PWL), born alive per litters Pathumthani 12120, Thailand (BAL), non-productive day (NPD) and wean to conception interval per litters (W2CL). A 3 Betagro Hybrid International Company Limited, total of 321 animals (140 Landrace and 181 Large White pigs) were genotyped with the Illumina Bangkok 10210, Thailand Porcine SNP 60k BeadChip, containing 61,177 single nucleotide polymorphisms (SNPs), ORCID while multiple traits single-step genomic BLUP method was used to calculate variances of Rattikan Suwannasing 5 SNP windows for 11,048 Landrace and 13,985 Large White data records. https://orcid.org/0000-0002-6950-4384 Monchai Duangjinda Results: The outcome of ssGWAS on the reproductive traits identified twenty-five and twenty- https://orcid.org/0000-0001-7044-8271 two SNPs associated with reproductive traits in Landrace and Large White, respectively. -

(NF1) As a Breast Cancer Driver
INVESTIGATION Comparative Oncogenomics Implicates the Neurofibromin 1 Gene (NF1) as a Breast Cancer Driver Marsha D. Wallace,*,† Adam D. Pfefferle,‡,§,1 Lishuang Shen,*,1 Adrian J. McNairn,* Ethan G. Cerami,** Barbara L. Fallon,* Vera D. Rinaldi,* Teresa L. Southard,*,†† Charles M. Perou,‡,§,‡‡ and John C. Schimenti*,†,§§,2 *Department of Biomedical Sciences, †Department of Molecular Biology and Genetics, ††Section of Anatomic Pathology, and §§Center for Vertebrate Genomics, Cornell University, Ithaca, New York 14853, ‡Department of Pathology and Laboratory Medicine, §Lineberger Comprehensive Cancer Center, and ‡‡Department of Genetics, University of North Carolina, Chapel Hill, North Carolina 27514, and **Memorial Sloan-Kettering Cancer Center, New York, New York 10065 ABSTRACT Identifying genomic alterations driving breast cancer is complicated by tumor diversity and genetic heterogeneity. Relevant mouse models are powerful for untangling this problem because such heterogeneity can be controlled. Inbred Chaos3 mice exhibit high levels of genomic instability leading to mammary tumors that have tumor gene expression profiles closely resembling mature human mammary luminal cell signatures. We genomically characterized mammary adenocarcinomas from these mice to identify cancer-causing genomic events that overlap common alterations in human breast cancer. Chaos3 tumors underwent recurrent copy number alterations (CNAs), particularly deletion of the RAS inhibitor Neurofibromin 1 (Nf1) in nearly all cases. These overlap with human CNAs including NF1, which is deleted or mutated in 27.7% of all breast carcinomas. Chaos3 mammary tumor cells exhibit RAS hyperactivation and increased sensitivity to RAS pathway inhibitors. These results indicate that spontaneous NF1 loss can drive breast cancer. This should be informative for treatment of the significant fraction of patients whose tumors bear NF1 mutations. -

Table S1. 103 Ferroptosis-Related Genes Retrieved from the Genecards
Table S1. 103 ferroptosis-related genes retrieved from the GeneCards. Gene Symbol Description Category GPX4 Glutathione Peroxidase 4 Protein Coding AIFM2 Apoptosis Inducing Factor Mitochondria Associated 2 Protein Coding TP53 Tumor Protein P53 Protein Coding ACSL4 Acyl-CoA Synthetase Long Chain Family Member 4 Protein Coding SLC7A11 Solute Carrier Family 7 Member 11 Protein Coding VDAC2 Voltage Dependent Anion Channel 2 Protein Coding VDAC3 Voltage Dependent Anion Channel 3 Protein Coding ATG5 Autophagy Related 5 Protein Coding ATG7 Autophagy Related 7 Protein Coding NCOA4 Nuclear Receptor Coactivator 4 Protein Coding HMOX1 Heme Oxygenase 1 Protein Coding SLC3A2 Solute Carrier Family 3 Member 2 Protein Coding ALOX15 Arachidonate 15-Lipoxygenase Protein Coding BECN1 Beclin 1 Protein Coding PRKAA1 Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 Protein Coding SAT1 Spermidine/Spermine N1-Acetyltransferase 1 Protein Coding NF2 Neurofibromin 2 Protein Coding YAP1 Yes1 Associated Transcriptional Regulator Protein Coding FTH1 Ferritin Heavy Chain 1 Protein Coding TF Transferrin Protein Coding TFRC Transferrin Receptor Protein Coding FTL Ferritin Light Chain Protein Coding CYBB Cytochrome B-245 Beta Chain Protein Coding GSS Glutathione Synthetase Protein Coding CP Ceruloplasmin Protein Coding PRNP Prion Protein Protein Coding SLC11A2 Solute Carrier Family 11 Member 2 Protein Coding SLC40A1 Solute Carrier Family 40 Member 1 Protein Coding STEAP3 STEAP3 Metalloreductase Protein Coding ACSL1 Acyl-CoA Synthetase Long Chain Family Member 1 Protein -
Single-Cell Transcriptome Analysis Reveals Mesenchymal Stem Cells In
bioRxiv preprint doi: https://doi.org/10.1101/2021.09.02.458742; this version posted September 3, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Single-cell transcriptome analysis reveals mesenchymal stem cells 2 in cavernous hemangioma 3 Fulong Ji1$, Yong Liu2$, Jinsong Shi3$, Chunxiang Liu1, Siqi Fu1 4 Heng Wang1, Bingbing Ren1, Dong Mi4, Shan Gao2*, Daqing Sun1* 5 1 Department of Paediatric Surgery, Tianjin Medical University General Hospital, Tianjin 300052, P.R. China. 6 China. 7 2 College of Life Sciences, Nankai University, Tianjin, Tianjin 300071, P.R. China; 8 3 National Clinical Research Center of Kidney Disease, Jinling Hospital, Nanjing University School of 9 Medicine, Nanjing, Jiangsu 210016, P.R. China; 10 4 School of Mathematical Sciences, Nankai University, Tianjin, Tianjin 300071, P.R. China; 11 12 13 $ These authors contributed equally to this paper. 14 * Corresponding authors. 15 SG:[email protected] 16 DS:[email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/2021.09.02.458742; this version posted September 3, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 A cavernous hemangioma, well-known as vascular malformation, is present at birth, grows 20 proportionately with the child, and does not undergo regression. -

Gene Modules Associated with Human Diseases Revealed by Network
bioRxiv preprint doi: https://doi.org/10.1101/598151; this version posted June 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Gene modules associated with human diseases revealed by network analysis Shisong Ma1,2*, Jiazhen Gong1†, Wanzhu Zuo1†, Haiying Geng1, Yu Zhang1, Meng Wang1, Ershang Han1, Jing Peng1, Yuzhou Wang1, Yifan Wang1, Yanyan Chen1 1. Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, China 2. School of Data Science, University of Science and Technology of China, Hefei, Anhui 230027, China * To whom correspondence should be addressed. Email: [email protected] † These authors contribute equally. 1 bioRxiv preprint doi: https://doi.org/10.1101/598151; this version posted June 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. ABSTRACT Despite many genes associated with human diseases have been identified, disease mechanisms often remain elusive due to the lack of understanding how disease genes are connected functionally at pathways level. Within biological networks, disease genes likely map to modules whose identification facilitates etiology studies but remains challenging. We describe a systematic approach to identify disease-associated gene modules. -

PDF Hosted at the Radboud Repository of the Radboud University Nijmegen
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/100868 Please be advised that this information was generated on 2017-12-06 and may be subject to change. Deciphering cellular responses to pathogens using genomics data Iziah Edwin Sama Deciphering cellular responses to pathogens using genomics data This research was performed at the Centre for Molecular and Biomolecular Informatics (CMBI), Nijmegen Centre of Molecular Life Sciences, Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands. Funding: This work was supported by the VIRGO consortium, an Innovative Cluster approved by the Netherlands Genomics Initiative and partially funded by the Dutch Government (BSIK 03012), The Netherlands. ISBN 978-90-9027062-3 © 2012 Iziah Edwin Sama All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, by print or otherwise, without permission in writing from the author Front Cover Image: A metaphorical illustration of the complexity in a host cell (the field), wherein fundamental moieties like proteins interact with each other (the network) in response to various pathogenic stimuli triggering respective cellular responses (the sub-fields demarcated by different line colors). The background is a picture of an indoors multi-sports field .The network is a protein-protein interaction network (HsapiensPPI of chapter 3) in which nodes represent proteins and edges between nodes indicate physical association. (Concept by Iziah Edwin Sama) Cover design and lay-out: In Zicht Grafisch Ontwerp, Arnhem Printed by: Ipskamp Drukkers, Enschede II Deciphering cellular responses to pathogens using genomics data Proefschrift ter verkrijging van de graad van doctor aan de Radboud Universiteit Nijmegen op gezag van de rector magnificus prof. -

Transcriptomic Profiling of Vascular Endothelial Growth Factor-Induced Signature Genes in Human Cervical Epithelial Cells A
TRANSCRIPTOMIC PROFILING OF VASCULAR ENDOTHELIAL GROWTH FACTOR-INDUCED SIGNATURE GENES IN HUMAN CERVICAL EPITHELIAL CELLS A Thesis by MACKINSEY DIANE JOHNSON Submitted to the Graduate School at Appalachian State University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 2019 Department of Biology TRANSCRIPTOMIC PROFILING OF VASCULAR ENDOTHELIAL GROWTH FACTOR-INDUCED SIGNATURE GENES IN HUMAN CERVICAL EPITHELIAL CELLS A Thesis by MACKINSEY DIANE JOHNSON August 2019 APPROVED BY: Chishimba Nathan Mowa, Ph.D. Chairperson, Thesis Committee Andrew Bellemer, Ph.D. Member, Thesis Committee Michael Opata, Ph.D. Member, Thesis Committee Zack E. Murrell, Ph.D. Chairperson, Department of Biology Michael J. McKenzie, Ph.D. Dean, Cratis D. Williams, School of Graduate Studies Copyright by MacKinsey Diane Johnson 2019 All Rights Reserved Abstract TRANSCRIPTOMIC PROFILING OF VASCULAR ENDOTHELIAL GROWTH FACTOR-INDUCED SIGNATURE GENES IN HUMAN CERVICAL EPITHELIAL CELLS MacKinsey Diane Johnson B.S., University of North Carolina at Chapel Hill Chairperson: Chishimba Nathan Mowa, Ph.D. Cervical epithelia cells play central roles in cervical remodeling (CR) during pregnancy and cervical events during the menstrual cycle, including mounting physical and immunological barriers, proliferation and differentiation, maintenance of fluid balance and likely in withstanding the mechanical force exerted by the growing fetus prior to term. We have previously characterized the cellular localization of vascular endothelial growth factor (VEGF), its receptor and signaling molecules in the cervix of rodents, its profile over the course of pregnancy and immediately after birth, as well as characterized its genome- and proteome-wide signature genes/proteins. These earlier studies reveal that VEGF and its associated molecules largely target and are localized in the cervical epithelial cells.