Transcriptomic Profiling of Vascular Endothelial Growth Factor-Induced Signature Genes in Human Cervical Epithelial Cells A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transcriptional Mechanisms of Resistance to Anti-PD-1 Therapy
Author Manuscript Published OnlineFirst on February 13, 2017; DOI: 10.1158/1078-0432.CCR-17-0270 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Transcriptional mechanisms of resistance to anti-PD-1 therapy Maria L. Ascierto1, Alvin Makohon-Moore2, 11, Evan J. Lipson1, Janis M. Taube3,4, Tracee L. McMiller5, Alan E. Berger6, Jinshui Fan6, Genevieve J. Kaunitz3, Tricia R. Cottrell4, Zachary A. Kohutek7, Alexander Favorov8,10, Vladimir Makarov7,11, Nadeem Riaz7,11, Timothy A. Chan7,11, Leslie Cope8, Ralph H. Hruban4,9, Drew M. Pardoll1, Barry S. Taylor11,12,13, David B. Solit13, Christine A Iacobuzio-Donahue2,11, and Suzanne L. Topalian5 From the 1Departments of Oncology, 3Dermatology, 4Pathology, 5Surgery, 6The Lowe Family Genomics Core, 8Oncology Bioinformatics Core, and the 9 Sol Goldman Pancreatic Cancer Research Center, Johns Hopkins University School of Medicine and Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD 21287; the 10Laboratory of System Biology and Computational Genetics, Vavilov Institute of General Genetics, Russian Academy of Sciences, 119991, Moscow, Russia; and 2Pathology, 7Radiation Oncology, 11Human Oncology and Pathogenesis Program, 12Department of Epidemiology and Biostatistics, and the 13Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York NY 10065. MLA, AM-M, EJL, and JMT contributed equally to this work Running title: Transcriptional mechanisms of resistance to anti-PD-1 Key Words: melanoma, cancer genetics, immunotherapy, anti-PD-1 Financial Support: This study was supported by the Melanoma Research Alliance (to SLT and CI-D), the Bloomberg~Kimmel Institute for Cancer Immunotherapy (to JMT, DMP, and SLT), the Barney Family Foundation (to SLT), Moving for Melanoma of Delaware (to SLT), the 1 Downloaded from clincancerres.aacrjournals.org on October 2, 2021. -

Environmental Influences on Endothelial Gene Expression
ENDOTHELIAL CELL GENE EXPRESSION John Matthew Jeff Herbert Supervisors: Prof. Roy Bicknell and Dr. Victoria Heath PhD thesis University of Birmingham August 2012 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Tumour angiogenesis is a vital process in the pathology of tumour development and metastasis. Targeting markers of tumour endothelium provide a means of targeted destruction of a tumours oxygen and nutrient supply via destruction of tumour vasculature, which in turn ultimately leads to beneficial consequences to patients. Although current anti -angiogenic and vascular targeting strategies help patients, more potently in combination with chemo therapy, there is still a need for more tumour endothelial marker discoveries as current treatments have cardiovascular and other side effects. For the first time, the analyses of in-vivo biotinylation of an embryonic system is performed to obtain putative vascular targets. Also for the first time, deep sequencing is applied to freshly isolated tumour and normal endothelial cells from lung, colon and bladder tissues for the identification of pan-vascular-targets. Integration of the proteomic, deep sequencing, public cDNA libraries and microarrays, delivers 5,892 putative vascular targets to the science community. -

Type of the Paper (Article
Cancers 2020, 12, S1 of S11 Supplementary Materials: Inflammatory Proteins HMGA2 and PRTN3 as Drivers of Vulvar Squamous Cell Carcinoma Pro- gression Agnieszka Fatalska, Natalia Rusetska, Elwira Bakuła-Zalewska, Artur Kowalik, Sebastian Zięba, Agnieszka Wroblewska, Kamil Zalewski, Krzysztof Goryca, Dominik Domański and Magdalena Kowalewska Text S1: Supplementary Methods iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) Cell Lysis The samples were snap-frozen in liquid nitrogen immediately after collection and stored at −70°C before pulverization (with the Microdismembrator II, B Braun Biotech In- ternational, Melsungen, Germany). Forty-two (14 control, 16 d-fVSCC and 12 progVSCC) samples were subjected to deep proteomic analysis. Total cell lysates were obtained from approximately 100 mg of pulverized patient tissue samples. The frozen pulverized tissue samples were resuspended in 300 µl of cold Lysis Buffer (1% sodium deoxycholate (NaDOC) in 25 mM HEPES (4-(2-hydroxyethyl)-1-pipera- zineethanesulfonic acid), boiled for 5 min at 100 °C, and cooled to room temperature (RT) on ice. Next, nucleic acids were degraded with 30 µL of benzonase nuclease (2.5 U/µL in 25 mM HEPES) at 4 °C for 30 min. Samples were then centrifuged twice at 12,000 × g for 10 min at 4 °C and the supernatants were stored at −80 °C. Protein concentrations were determined using the Pierce (Appleton, WI, USA) Bicinchoninic Acid (BCA) Assay Kit according to the manufacturer’s instructions, for the 96-well plate format, in duplicate, at two different sample dilutions (ThermoFisher Scientific, Waltham, MA, USA). Sample Protein Extract Digestion and iTRAQ Labeling Protein extract digestion and iTRAQ tag labeling was conducted using the 8-plex iTRAQ assay and iTRAQ Reagent and Buffer Kits (AB SCIEX, Framingham, MA, USA). -
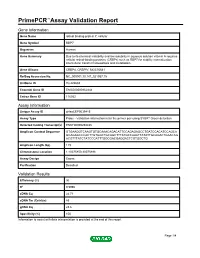
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name retinol binding protein 7, cellular Gene Symbol RBP7 Organism Human Gene Summary Due to its chemical instability and low solubility in aqueous solution vitamin A requires cellular retinol-binding proteins (CRBPs) such as RBP7 for stability internalization intercellular transfer homeostasis and metabolism. Gene Aliases CRBP4, CRBPIV, MGC70641 RefSeq Accession No. NC_000001.10, NT_021937.19 UniGene ID Hs.422688 Ensembl Gene ID ENSG00000162444 Entrez Gene ID 116362 Assay Information Unique Assay ID qHsaCEP0039415 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000294435 Amplicon Context Sequence GTGAAGGTCAAGTGTGCAAACAGACATTCCAGAGAGCCTGATCCACATCCAGCA GCAGAGCCCACTTGTGGCTGCAGCTTTATGCCAAATTATATTGCAGACTGAACAG ACGTTTATCTATCCCATTTGGCGACGAGGACTCGTGGCTG Amplicon Length (bp) 119 Chromosome Location 1:10075850-10075998 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 96 R2 0.9996 cDNA Cq 24.71 cDNA Tm (Celsius) 83 gDNA Cq 23.6 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report RBP7, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Homeobox Genes Gain Trimethylation of Histone H3 Lysine 4 in Glioblastoma Tissue
Biosci. Rep. (2016) / 36 / art:e00347 / doi 10.1042/BSR20160028 Homeobox genes gain trimethylation of histone H3 lysine 4 in glioblastoma tissue Kun Luo*1, Donghui Luo† and Hao Wen‡1 *Department of Neurosurgery, Xinjiang Evidence-Based Medicine Research Institute, First Affiliated Hospital of Xinjiang Medical University, Urumqi 830054, China †Department of Neurology, First Affiliated Hospital of Xinjiang Medical University, Urumqi 830054, China ‡State Key Laboratory Incubation Base of Xinjiang Major Diseases Research, First Affiliated Hospital of Xinjiang Medical University, Urumqi 830054, China Synopsis Glioblastoma multiforme (GBM) exhibits considerable heterogeneity and associates with genome-wide alterations of the repressed chromatin marks DNA methylation and H3 lysine 27 trimethylation (H3K27me3). Tri-methylation on lysine 4 of histone H3 (H3K4me3) is an activating epigenetic mark that is enriched at promoter and promotes expression. It will be helpful in GBM diagnosis and treatment to identify the alteration of H3K4me3 between human GBM and GBM-surrounding tissues. Here, we performed an analysis using next-generation sequencing techniques to identify H3K4me3 modification in a case of GBM and the GBM-surrounding tissues. The results revealed a global decrease in H3K4me3 in GBM, especially at promoters and CpG islands. In GBM, homeobox genes gain H3K4me3, whereas the cell–cell adhesion-related cadherin genes lose H3K4me3. The products of the homeobox genes are highly connected with Ras-signalling and PI3K-Akt signalling pathways. Using The Cancer Genome Atlas (TCGA) data, we inferred the homeobox-regulated genes’ expression is higher in 548 GBM cases than in 27 lower grade glioma cases giving that OLIG2 expression can be a reference. -

PCDHA3 Antibody (C-Term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap12144b
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 PCDHA3 Antibody (C-term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP12144b Specification PCDHA3 Antibody (C-term) - Product Information Application WB, IHC-P,E Primary Accession Q9Y5H8 Other Accession NP_061729.1, NP_113685.1 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Calculated MW 102428 Antigen Region 773-800 PCDHA3 Antibody (C-term) - Additional Information PCDHA3 Antibody (C-term) (Cat. #AP12144b) western blot analysis in Gene ID 56145 MDA-MB231 cell line lysates (35ug/lane).This Other Names demonstrates the PCDHA3 antibody detected Protocadherin alpha-3, PCDH-alpha-3, the PCDHA3 protein (arrow). PCDHA3 Target/Specificity This PCDHA3 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 773-800 amino acids from the C-terminal region of human PCDHA3. Dilution WB~~1:1000 IHC-P~~1:10~50 Format Purified polyclonal antibody supplied in PBS PCDHA3 Antibody (C-term) (Cat. with 0.09% (W/V) sodium azide. This #AP12144b)immunohistochemistry analysis antibody is purified through a protein A in formalin fixed and paraffin embedded column, followed by peptide affinity human brain tissue followed by peroxidase purification. conjugation of the secondary antibody and DAB staining.This data demonstrates the use Storage of PCDHA3 Antibody (C-term) for Maintain refrigerated at 2-8°C for up to 2 immunohistochemistry. Clinical relevance has weeks. For long term storage store at -20°C not been evaluated. in small aliquots to prevent freeze-thaw cycles. PCDHA3 Antibody (C-term) - Background Precautions Page 1/2 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 PCDHA3 Antibody (C-term) is for research This gene is a member of the protocadherin use only and not for use in diagnostic or alpha gene therapeutic procedures. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Acetyl-Histone H2A-K5 Rabbit Pab
Leader in Biomolecular Solutions for Life Science Acetyl-Histone H2A-K5 Rabbit pAb Catalog No.: A15620 Basic Information Background Catalog No. Histones are basic nuclear proteins that are responsible for the nucleosome structure of A15620 the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is Observed MW wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with 14kDa linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene is intronless and encodes a replication-dependent Calculated MW histone that is a member of the histone H2A family. Transcripts from this gene lack polyA 14kDa tails but instead contain a palindromic termination element. This gene is found in the small histone gene cluster on chromosome 6p22-p21.3. Category Primary antibody Applications WB, IHC, IF Cross-Reactivity Human, Mouse, Rat, Other (Wide Range) Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 8329 P0C0S8 IHC 1:50 - 1:100 Immunogen IF 1:50 - 1:100 A synthetic acetylated peptide around K5 of human Histone H2A (NP_003508.1). Synonyms HIST1H2AI;H2A/c;H2AFC Contact Product Information Source Isotype Purification www.abclonal.com Rabbit IgG Affinity purification Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of various cell lines, using Acetyl-Histone H2A-K5 antibody (A15620) at 1:1000 dilution.C2C12 cells and C6 cells were treated by TSA (1 uM) at 37℃ for 18 hours. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Regulation of RNA Interference Pathways in the Insect Vector Laodelphax Striatellus by Viral Proteins of Rice Stripe Virus
viruses Article Regulation of RNA Interference Pathways in the Insect Vector Laodelphax striatellus by Viral Proteins of Rice Stripe Virus Yan Xiao 1,2,†, Qiong Li 1,3,†, Wei Wang 1, Yumei Fu 4 and Feng Cui 1,3,* 1 State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; [email protected] (Y.X.); [email protected] (Q.L.); [email protected] (W.W.) 2 College of Life Sciences, Hebei University, Baoding 071002, China 3 CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing 100049, China 4 Key Laboratory of Tropical Translational Medicine of Ministry of Education, School of Tropical Medicine and Laboratory Medicine, Hainan Medical University, Haikou 571199, China; [email protected] * Correspondence: [email protected] † These authors contributed equally to this work. Abstract: RNA interference (RNAi), especially the small interfering RNA (siRNA) and microRNA (miRNA) pathways, plays an important role in defending against viruses in plants and insects. However, how insect-transmitted phytoviruses regulate the RNAi-mediated antiviral response in vector insects has barely been uncovered. In this study, we explored the interaction between rice stripe virus (RSV) and the miRNA and siRNA pathways of the small brown planthopper, which is a vector insect. The transcript and protein levels of key genes in the two RNAi pathways did not change during the RSV infection process. When the expression of insect Ago1, Ago2, or Translin was silenced by the injection of double-stranded RNAs targeting these genes, viral replication was Citation: Xiao, Y.; Li, Q.; Wang, W.; promoted with Ago2 silencing but inhibited with Translin silencing. -

Whole Exome Sequencing in Families at High Risk for Hodgkin Lymphoma: Identification of a Predisposing Mutation in the KDR Gene
Hodgkin Lymphoma SUPPLEMENTARY APPENDIX Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene Melissa Rotunno, 1 Mary L. McMaster, 1 Joseph Boland, 2 Sara Bass, 2 Xijun Zhang, 2 Laurie Burdett, 2 Belynda Hicks, 2 Sarangan Ravichandran, 3 Brian T. Luke, 3 Meredith Yeager, 2 Laura Fontaine, 4 Paula L. Hyland, 1 Alisa M. Goldstein, 1 NCI DCEG Cancer Sequencing Working Group, NCI DCEG Cancer Genomics Research Laboratory, Stephen J. Chanock, 5 Neil E. Caporaso, 1 Margaret A. Tucker, 6 and Lynn R. Goldin 1 1Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 2Cancer Genomics Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 3Ad - vanced Biomedical Computing Center, Leidos Biomedical Research Inc.; Frederick National Laboratory for Cancer Research, Frederick, MD; 4Westat, Inc., Rockville MD; 5Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; and 6Human Genetics Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA ©2016 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2015.135475 Received: August 19, 2015. Accepted: January 7, 2016. Pre-published: June 13, 2016. Correspondence: [email protected] Supplemental Author Information: NCI DCEG Cancer Sequencing Working Group: Mark H. Greene, Allan Hildesheim, Nan Hu, Maria Theresa Landi, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Anand Pathak, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, Xiaohong R. Yang, Guoqin Yu NCI DCEG Cancer Genomics Research Laboratory: Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A. -

Hippo and Sonic Hedgehog Signalling Pathway Modulation of Human Urothelial Tissue Homeostasis
Hippo and Sonic Hedgehog signalling pathway modulation of human urothelial tissue homeostasis Thomas Crighton PhD University of York Department of Biology November 2020 Abstract The urinary tract is lined by a barrier-forming, mitotically-quiescent urothelium, which retains the ability to regenerate following injury. Regulation of tissue homeostasis by Hippo and Sonic Hedgehog signalling has previously been implicated in various mammalian epithelia, but limited evidence exists as to their role in adult human urothelial physiology. Focussing on the Hippo pathway, the aims of this thesis were to characterise expression of said pathways in urothelium, determine what role the pathways have in regulating urothelial phenotype, and investigate whether the pathways are implicated in muscle-invasive bladder cancer (MIBC). These aims were assessed using a cell culture paradigm of Normal Human Urothelial (NHU) cells that can be manipulated in vitro to represent different differentiated phenotypes, alongside MIBC cell lines and The Cancer Genome Atlas resource. Transcriptomic analysis of NHU cells identified a significant induction of VGLL1, a poorly understood regulator of Hippo signalling, in differentiated cells. Activation of upstream transcription factors PPARγ and GATA3 and/or blockade of active EGFR/RAS/RAF/MEK/ERK signalling were identified as mechanisms which induce VGLL1 expression in NHU cells. Ectopic overexpression of VGLL1 in undifferentiated NHU cells and MIBC cell line T24 resulted in significantly reduced proliferation. Conversely, knockdown of VGLL1 in differentiated NHU cells significantly reduced barrier tightness in an unwounded state, while inhibiting regeneration and increasing cell cycle activation in scratch-wounded cultures. A signalling pathway previously observed to be inhibited by VGLL1 function, YAP/TAZ, was unaffected by VGLL1 manipulation.