The Concise Guide to PHARMACOLOGY 2013/14: Overview
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Selectivity and Partial Efficacy at the Monoamine Transporters: a Unified Model of Allosteric Modulation and Amphetamine-Induced Substrate Release
1521-0111/95/3/303–312$35.00 https://doi.org/10.1124/mol.118.114793 MOLECULAR PHARMACOLOGY Mol Pharmacol 95:303–312, March 2019 Copyright ª 2019 by The Author(s) This is an open access article distributed under the CC BY-NC Attribution 4.0 International license. MINIREVIEW Functional Selectivity and Partial Efficacy at the Monoamine Transporters: A Unified Model of Allosteric Modulation and Amphetamine-Induced Substrate Release Peter S. Hasenhuetl,1,2 Shreyas Bhat,2 Michael Freissmuth, and Walter Sandtner Downloaded from Institute of Pharmacology, Gaston H. Glock Research Laboratories for Exploratory Drug Development, Center of Physiology and Pharmacology, Medical University of Vienna, Vienna, Austria Received October 2, 2018; accepted December 13, 2018 ABSTRACT molpharm.aspetjournals.org All clinically approved drugs targeting the plasmalemmal trans- allosteric inhibition and stimulation of MATs, which can be porters for dopamine, norepinephrine, and serotonin act either functionally selective for either substrate uptake or amphetamine- as competitive uptake inhibitors or as amphetamine-like releas- induced release. These concepts are integrated into a parsimonious ers. Monoamine transporter (MAT) ligands that allosterically kinetic framework, which relies exclusively on physiologic transport affect MAT-mediated substrate uptake, release, or both were modes (without any deviation from an alternating access mecha- recently discovered. Their modes of action have not yet been nism). The model posits cooperative substrate and Na1 binding and explained in a unified framework. Here, we go beyond compet- functional selectivity by conformational selection (i.e., preference of itive inhibitors and classic amphetamines and introduce con- the allosteric modulators for the substrate-loaded or substrate-free cepts for partial efficacy at and allosteric modulation of MATs. -

Ageing, Sex and Cardioprotection Marisol Ruiz-Meana1,2, Kerstin Boengler3, David Garcia-Dorado1,2, Derek J
Kaambre Tuuli (Orcid ID: 0000-0001-5755-4694) Kararigas Georgios (Orcid ID: 0000-0002-8187-0176) Ageing, sex and cardioprotection Marisol Ruiz-Meana1,2, Kerstin Boengler3, David Garcia-Dorado1,2, Derek J. Hausenloy4,5,6,7,8,9, Tuuli Kaambre10, Georgios Kararigas11,12, Cinzia Perrino13, Rainer Schulz3, Kirsti Ytrehus14. 1 Hospital Universitari Vall d’Hebron, Department of Cardiology. Vall d’Hebron Institut de Recerca (VHIR).Universitat Autonoma de Barcelona, Spain. 2 Centro de Investigación Biomédica en Red-CV, CIBER-CV, Spain. 3Institute of Physiology, Justus-Liebig University Giessen. Giessen 35392, Aulweg 129, Germany. 4Cardiovascular & Metabolic Disorders Program, Duke-National University of Singapore Medical School, Singapore. 5National Heart Research Institute Singapore, National Heart Centre, Singapore. 6Yong Loo Lin School of Medicine, National University Singapore, Singapore. 7The Hatter Cardiovascular Institute, University College London, London, UK. 8The National Institute of Health Research University College London Hospitals Biomedical Research Centre, Research & Development, London, UK. 9Tecnologico de Monterrey, Centro de Biotecnologia-FEMSA, Nuevo Leon, Mexico. 10Laboratory of Chemical Biology, National Institute of Chemical Physics and Biophysics, Akadeemia tee 23, 12618 Tallinn, Estonia. 11Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin, Germany. 12DZHK (German Centre for Cardiovascular Research), partner site Berlin, -
Primepcr™Assay Validation Report
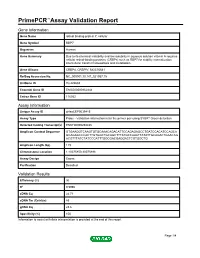
PrimePCR™Assay Validation Report Gene Information Gene Name retinol binding protein 7, cellular Gene Symbol RBP7 Organism Human Gene Summary Due to its chemical instability and low solubility in aqueous solution vitamin A requires cellular retinol-binding proteins (CRBPs) such as RBP7 for stability internalization intercellular transfer homeostasis and metabolism. Gene Aliases CRBP4, CRBPIV, MGC70641 RefSeq Accession No. NC_000001.10, NT_021937.19 UniGene ID Hs.422688 Ensembl Gene ID ENSG00000162444 Entrez Gene ID 116362 Assay Information Unique Assay ID qHsaCEP0039415 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000294435 Amplicon Context Sequence GTGAAGGTCAAGTGTGCAAACAGACATTCCAGAGAGCCTGATCCACATCCAGCA GCAGAGCCCACTTGTGGCTGCAGCTTTATGCCAAATTATATTGCAGACTGAACAG ACGTTTATCTATCCCATTTGGCGACGAGGACTCGTGGCTG Amplicon Length (bp) 119 Chromosome Location 1:10075850-10075998 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 96 R2 0.9996 cDNA Cq 24.71 cDNA Tm (Celsius) 83 gDNA Cq 23.6 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report RBP7, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Full-Text PDF (Final Published Version)
Alexander, S. P. H., Cidlowski, J. A., Kelly, E., Marrion, N. V., Peters, J. A., Faccenda, E., Harding, S. D., Pawson, A. J., Sharman, J. L., Southan, C., Davies, J. A., & CGTP Collaborators (2017). THE CONCISE GUIDE TO PHARMACOLOGY 2017/18: Nuclear hormone receptors. British Journal of Pharmacology, 174, S208-S224. https://doi.org/10.1111/bph.13880 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1111/bph.13880 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via Wiley at https://doi.org/10.1111/bph.13880 . Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2017/18: Nuclear hormone receptors. British Journal of Pharmacology (2017) 174, S208–S224 THE CONCISE GUIDE TO PHARMACOLOGY 2017/18: Nuclear hormone receptors Stephen PH Alexander1, John A Cidlowski2, Eamonn Kelly3, Neil V Marrion3, John A Peters4, Elena Faccenda5, Simon D Harding5,AdamJPawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical -

Insights Into Nuclear G-Protein-Coupled Receptors As Therapeutic Targets in Non-Communicable Diseases
pharmaceuticals Review Insights into Nuclear G-Protein-Coupled Receptors as Therapeutic Targets in Non-Communicable Diseases Salomé Gonçalves-Monteiro 1,2, Rita Ribeiro-Oliveira 1,2, Maria Sofia Vieira-Rocha 1,2, Martin Vojtek 1,2 , Joana B. Sousa 1,2,* and Carmen Diniz 1,2,* 1 Laboratory of Pharmacology, Department of Drug Sciences, Faculty of Pharmacy, University of Porto, 4050-313 Porto, Portugal; [email protected] (S.G.-M.); [email protected] (R.R.-O.); [email protected] (M.S.V.-R.); [email protected] (M.V.) 2 LAQV/REQUIMTE, Faculty of Pharmacy, University of Porto, 4050-313 Porto, Portugal * Correspondence: [email protected] (J.B.S.); [email protected] (C.D.) Abstract: G-protein-coupled receptors (GPCRs) comprise a large protein superfamily divided into six classes, rhodopsin-like (A), secretin receptor family (B), metabotropic glutamate (C), fungal mating pheromone receptors (D), cyclic AMP receptors (E) and frizzled (F). Until recently, GPCRs signaling was thought to emanate exclusively from the plasma membrane as a response to extracellular stimuli but several studies have challenged this view demonstrating that GPCRs can be present in intracellular localizations, including in the nuclei. A renewed interest in GPCR receptors’ superfamily emerged and intensive research occurred over recent decades, particularly regarding class A GPCRs, but some class B and C have also been explored. Nuclear GPCRs proved to be functional and capable of triggering identical and/or distinct signaling pathways associated with their counterparts on the cell surface bringing new insights into the relevance of nuclear GPCRs and highlighting the Citation: Gonçalves-Monteiro, S.; nucleus as an autonomous signaling organelle (triggered by GPCRs). -

Modulatory Roles of ATP and Adenosine in Cholinergic Neuromuscular Transmission
International Journal of Molecular Sciences Review Modulatory Roles of ATP and Adenosine in Cholinergic Neuromuscular Transmission Ayrat U. Ziganshin 1,* , Adel E. Khairullin 2, Charles H. V. Hoyle 1 and Sergey N. Grishin 3 1 Department of Pharmacology, Kazan State Medical University, 49 Butlerov Street, 420012 Kazan, Russia; [email protected] 2 Department of Biochemistry, Laboratory and Clinical Diagnostics, Kazan State Medical University, 49 Butlerov Street, 420012 Kazan, Russia; [email protected] 3 Department of Medical and Biological Physics with Computer Science and Medical Equipment, Kazan State Medical University, 49 Butlerov Street, 420012 Kazan, Russia; [email protected] * Correspondence: [email protected]; Tel.: +7-843-236-0512 Received: 30 June 2020; Accepted: 1 September 2020; Published: 3 September 2020 Abstract: A review of the data on the modulatory action of adenosine 5’-triphosphate (ATP), the main co-transmitter with acetylcholine, and adenosine, the final ATP metabolite in the synaptic cleft, on neuromuscular transmission is presented. The effects of these endogenous modulators on pre- and post-synaptic processes are discussed. The contribution of purines to the processes of quantal and non- quantal secretion of acetylcholine into the synaptic cleft, as well as the influence of the postsynaptic effects of ATP and adenosine on the functioning of cholinergic receptors, are evaluated. As usual, the P2-receptor-mediated influence is minimal under physiological conditions, but it becomes very important in some pathophysiological situations such as hypothermia, stress, or ischemia. There are some data demonstrating the same in neuromuscular transmission. It is suggested that the role of endogenous purines is primarily to provide a safety factor for the efficiency of cholinergic neuromuscular transmission. -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Vertebrate Fatty Acid and Retinoid Binding Protein Genes and Proteins: Evidence for Ancient and Recent Gene Duplication Events
In: Advances in Genetics Research. Volume 11 ISBN: 978-1-62948-744-1 Editor: Kevin V. Urbano © 2014 Nova Science Publishers, Inc. Chapter 7 Vertebrate Fatty Acid and Retinoid Binding Protein Genes and Proteins: Evidence for Ancient and Recent Gene Duplication Events Roger S. Holmes Eskitis Institute for Drug Discovery and School of Biomolecular and Physical Sciences, Griffith University, Nathan, QLD, Australia Abstract Fatty acid binding proteins (FABP) and retinoid binding proteins (RBP) are members of a family of small, highly conserved cytoplasmic proteins that function in binding and facilitating the cellular uptake of fatty acids, retinoids and other hydrophobic compounds. Several human FABP-like genes are expressed in the body: FABP1 (liver); FABP2 (intestine); FABP3 (heart and skeletal muscle); FABP4 (adipocyte); FABP5 (epidermis); FABP6 (ileum); FABP7 (brain); FABP8 (nervous system); FABP9 (testis); and FABP12 (retina and testis). A related gene (FABP10) is expressed in lower vertebrate liver and other tissues. Four RBP genes are expressed in human tissues: RBP1 (many tissues); RBP2 (small intestine epithelium); RBP5 (kidney and liver); and RBP7 (kidney and heart). Comparative FABP and RBP amino acid sequences and structures and gene locations were examined using data from several vertebrate genome projects. Sequence alignments, key amino acid residues and conserved predicted secondary and tertiary structures were also studied, including lipid binding regions. Vertebrate FABP- and RBP- like genes usually contained 4 coding exons in conserved locations, supporting a common evolutionary origin for these genes. Phylogenetic analyses examined the relationships and evolutionary origins of these genes, suggesting division into three FABP gene classes: 1: FABP1, FABP6 and FABP10; 2: FABP2; and 3, with 2 groups: 3A: FABP4, FABP8, FABP9 and FABP12; and 3B: and FABP3, FABP5 and FABP7. -

NADPH Oxidases (Version 2019.4) in the IUPHAR/BPS Guide to Pharmacology Database
IUPHAR/BPS Guide to Pharmacology CITE https://doi.org/10.2218/gtopdb/F993/2019.4 NADPH oxidases (version 2019.4) in the IUPHAR/BPS Guide to Pharmacology Database Albert van der Vliet1 1. University of Vermont, USA Abstract The two DUOX enzymes were originally identified as participating in the production of hydrogen peroxide as a pre-requisite for thyroid hormone biosynthesis in the thyroid gland [6]. NOX enzymes function to catalyse the reduction of molecular oxygen to superoxide and various other reactive oxygen species (ROS). They are subunits of the NADPH oxidase complex. Contents This is a citation summary for NADPH oxidases in the Guide to Pharmacology database (GtoPdb). It exists purely as an adjunct to the database to facilitate the recognition of citations to and from the database by citation analyzers. Readers will almost certainly want to visit the relevant sections of the database which are given here under database links. GtoPdb is an expert-driven guide to pharmacological targets and the substances that act on them. GtoPdb is a reference work which is most usefully represented as an on-line database. As in any publication this work should be appropriately cited, and the papers it cites should also be recognized. This document provides a citation for the relevant parts of the database, and also provides a reference list for the research cited by those parts. Please note that the database version for the citations given in GtoPdb are to the most recent preceding version in which the family or its subfamilies and targets were substantially changed. The links below are to the current version. -

Sex Steroids Regulate Skin Pigmentation Through Nonclassical
RESEARCH ARTICLE Sex steroids regulate skin pigmentation through nonclassical membrane-bound receptors Christopher A Natale1, Elizabeth K Duperret1, Junqian Zhang1, Rochelle Sadeghi1, Ankit Dahal1, Kevin Tyler O’Brien2, Rosa Cookson2, Jeffrey D Winkler2, Todd W Ridky1* 1Department of Dermatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, United States; 2Department of Chemistry, University of Pennsylvania, Philadelphia, United States Abstract The association between pregnancy and altered cutaneous pigmentation has been documented for over two millennia, suggesting that sex hormones play a role in regulating epidermal melanocyte (MC) homeostasis. Here we show that physiologic estrogen (17b-estradiol) and progesterone reciprocally regulate melanin synthesis. This is intriguing given that we also show that normal primary human MCs lack classical estrogen or progesterone receptors (ER or PR). Utilizing both genetic and pharmacologic approaches, we establish that sex steroid effects on human pigment synthesis are mediated by the membrane-bound, steroid hormone receptors G protein-coupled estrogen receptor (GPER), and progestin and adipoQ receptor 7 (PAQR7). Activity of these receptors was activated or inhibited by synthetic estrogen or progesterone analogs that do not bind to ER or PR. As safe and effective treatment options for skin pigmentation disorders are limited, these specific GPER and PAQR7 ligands may represent a novel class of therapeutics. DOI: 10.7554/eLife.15104.001 *For correspondence: ridky@mail. -

Hyperlinking from BJP Articles to the Guide to PHARMACOLOGY
Hyperlinking from BJP articles to the Guide to PHARMACOLOGY BJP requires the authors of articles accepted for publication to hyperlink the key protein targets and ligands in their article to the corresponding entries in the IUPHAR/BPS Guide to PHARMACOLOGY (http://www.guidetopharmacology.org), as shown in the example below: Introduction GABAA-ρ as designated by International Union of Pharmacology (IUPHAR) (Alexander et al., 2015) and also known as GABA-ρ or GABAC receptors, are homopentameric ligand-gated ion channels (LGIC) composed of ρ subunits. They are members of the pentameric or Cys-loop LGIC superfamily comprising excitatory cation selective receptors such as nicotinic acetylcholine receptors, 5-HT3 receptors and zinc-activated channels, and inhibitory anion-selective receptors such as GABAA receptors, strychnine-sensitive glycine receptors and invertebrate glutamate- gated chloride channels (Thompson et al., 2010; Baenziger and Corringer, 2011). Receptors of this superfamily require five subunits to assemble a single ion channel. The ion channel may be homomeric formed by five identical subunits as is the case of GABA-ρ receptors, or heteromeric, consisting of a combination of at least two different subunits, such as GABAA receptors (Olsen and Sieghart, 2009). Hyperlinks Only key targets and ligands should be hyperlinked, and they should only be hyperlinked at the first mention of them in the main text. Please also create a list of the hyperlinks applied to your article in a separate word file. Please choose ‘Data Files’ as the File Designation when you upload the word file and ensure the name of the file is ‘List of Hyperlinks for Crosschecking’. -

Transporters
Alexander, S. P. H., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., Armstrong, J. F., Faccenda, E., Harding, S. D., Pawson, A. J., Sharman, J. L., Southan, C., Davies, J. A., & CGTP Collaborators (2019). The Concise Guide to Pharmacology 2019/20: Transporters. British Journal of Pharmacology, 176(S1), S397-S493. https://doi.org/10.1111/bph.14753 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1111/bph.14753 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via Wiley at https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14753. Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L Sharman5 , Christopher Southan5 , Jamie A Davies5 and CGTP Collaborators 1School of Life Sciences,