Cancer Informatics 2014:13 13–20 Doi: 10.4137/CIN.S13495
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
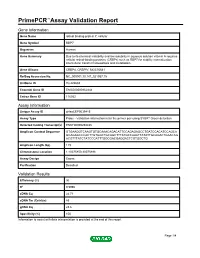
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name retinol binding protein 7, cellular Gene Symbol RBP7 Organism Human Gene Summary Due to its chemical instability and low solubility in aqueous solution vitamin A requires cellular retinol-binding proteins (CRBPs) such as RBP7 for stability internalization intercellular transfer homeostasis and metabolism. Gene Aliases CRBP4, CRBPIV, MGC70641 RefSeq Accession No. NC_000001.10, NT_021937.19 UniGene ID Hs.422688 Ensembl Gene ID ENSG00000162444 Entrez Gene ID 116362 Assay Information Unique Assay ID qHsaCEP0039415 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000294435 Amplicon Context Sequence GTGAAGGTCAAGTGTGCAAACAGACATTCCAGAGAGCCTGATCCACATCCAGCA GCAGAGCCCACTTGTGGCTGCAGCTTTATGCCAAATTATATTGCAGACTGAACAG ACGTTTATCTATCCCATTTGGCGACGAGGACTCGTGGCTG Amplicon Length (bp) 119 Chromosome Location 1:10075850-10075998 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 96 R2 0.9996 cDNA Cq 24.71 cDNA Tm (Celsius) 83 gDNA Cq 23.6 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report RBP7, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Epigenome-Wide Exploratory Study of Monozygotic Twins Suggests Differentially Methylated Regions to Associate with Hand Grip Strength
Biogerontology (2019) 20:627–647 https://doi.org/10.1007/s10522-019-09818-1 (0123456789().,-volV)( 0123456789().,-volV) RESEARCH ARTICLE Epigenome-wide exploratory study of monozygotic twins suggests differentially methylated regions to associate with hand grip strength Mette Soerensen . Weilong Li . Birgit Debrabant . Marianne Nygaard . Jonas Mengel-From . Morten Frost . Kaare Christensen . Lene Christiansen . Qihua Tan Received: 15 April 2019 / Accepted: 24 June 2019 / Published online: 28 June 2019 Ó The Author(s) 2019 Abstract Hand grip strength is a measure of mus- significant CpG sites or pathways were found, how- cular strength and is used to study age-related loss of ever two of the suggestive top CpG sites were mapped physical capacity. In order to explore the biological to the COL6A1 and CACNA1B genes, known to be mechanisms that influence hand grip strength varia- related to muscular dysfunction. By investigating tion, an epigenome-wide association study (EWAS) of genomic regions using the comb-p algorithm, several hand grip strength in 672 middle-aged and elderly differentially methylated regions in regulatory monozygotic twins (age 55–90 years) was performed, domains were identified as significantly associated to using both individual and twin pair level analyses, the hand grip strength, and pathway analyses of these latter controlling the influence of genetic variation. regions revealed significant pathways related to the Moreover, as measurements of hand grip strength immune system, autoimmune disorders, including performed over 8 years were available in the elderly diabetes type 1 and viral myocarditis, as well as twins (age 73–90 at intake), a longitudinal EWAS was negative regulation of cell differentiation. -

Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’S Biological Network Based on Systematic Pharmacology
ORIGINAL RESEARCH published: 25 June 2021 doi: 10.3389/fphar.2021.601846 Exploring the Regulatory Mechanism of Hedysarum Multijugum Maxim.-Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’s Biological Network Based on Systematic Pharmacology Kailin Yang 1†, Liuting Zeng 1†, Anqi Ge 2†, Yi Chen 1†, Shanshan Wang 1†, Xiaofei Zhu 1,3† and Jinwen Ge 1,4* Edited by: 1 Takashi Sato, Key Laboratory of Hunan Province for Integrated Traditional Chinese and Western Medicine on Prevention and Treatment of 2 Tokyo University of Pharmacy and Life Cardio-Cerebral Diseases, Hunan University of Chinese Medicine, Changsha, China, Galactophore Department, The First 3 Sciences, Japan Hospital of Hunan University of Chinese Medicine, Changsha, China, School of Graduate, Central South University, Changsha, China, 4Shaoyang University, Shaoyang, China Reviewed by: Hui Zhao, Capital Medical University, China Background: Clinical research found that Hedysarum Multijugum Maxim.-Chuanxiong Maria Luisa Del Moral, fi University of Jaén, Spain Rhizoma Compound (HCC) has de nite curative effect on cerebral ischemic diseases, *Correspondence: such as ischemic stroke and cerebral ischemia-reperfusion injury (CIR). However, its Jinwen Ge mechanism for treating cerebral ischemia is still not fully explained. [email protected] †These authors share first authorship Methods: The traditional Chinese medicine related database were utilized to obtain the components of HCC. The Pharmmapper were used to predict HCC’s potential targets. Specialty section: The CIR genes were obtained from Genecards and OMIM and the protein-protein This article was submitted to interaction (PPI) data of HCC’s targets and IS genes were obtained from String Ethnopharmacology, a section of the journal database. -

Supreme Court of the United States
No. 12-398 IN THE Supreme Court of the United States THE ASSOCIATION FOR MOLECULAR PATHOLOGY, ET AL., Petitioners, v. MYRIAD GENETICS, INC., ET AL., Respondents. __________ On Writ of Certiorari to the United States Court of Appeals for the Federal Circuit __________ BRIEF OF AMICI CURIAE AMERICAN MEDICAL ASSOCIATION, AMERICAN SOCIETY OF HUMAN GENETICS, AMERICAN COLLEGE OF OBSTETRICIANS AND GYNECOLOGISTS, AMERICAN OSTEOPATHIC ASSOCIATION, AMERICAN COLLEGE OF LEGAL MEDICINE, AND THE MEDICAL SOCIETY OF THE STATE OF NEW YORK IN SUPPORT OF PETITIONERS __________ Professor Lori B. Andrews Counsel of Record Chicago-Kent College of Law Illinois Institute of Technology 565 West Adams Street Chicago, IL 60661 [email protected] 312-906-5359 TABLE OF CONTENTS TABLE OF AUTHORITIES ......................................... iii STATEMENT OF INTEREST OF AMICI CURIAE .... 1 SUMMARY OF THE ARGUMENT .............................. 4 ARGUMENT .................................................................. 6 I. PHYSICIANS’ AND RESEARCHERS’ ACCESS TO HUMAN GENE SEQUENCES IS VITAL TO HEALTH CARE AND RESEARCH ........................................................ 6 A. Patents on Human Genes Interfere with Diagnosis and Treatment of Patients ..... 7 B. Patents on Human Genes Increase the Cost of Genetic Testing .......................... 11 C. Patents on Human Genes Impede Innovation .............................................. 13 D. Existing Non-Patent Incentives Are Sufficient to Encourage Innovation in Genetics ................................................. -

Hippo and Sonic Hedgehog Signalling Pathway Modulation of Human Urothelial Tissue Homeostasis
Hippo and Sonic Hedgehog signalling pathway modulation of human urothelial tissue homeostasis Thomas Crighton PhD University of York Department of Biology November 2020 Abstract The urinary tract is lined by a barrier-forming, mitotically-quiescent urothelium, which retains the ability to regenerate following injury. Regulation of tissue homeostasis by Hippo and Sonic Hedgehog signalling has previously been implicated in various mammalian epithelia, but limited evidence exists as to their role in adult human urothelial physiology. Focussing on the Hippo pathway, the aims of this thesis were to characterise expression of said pathways in urothelium, determine what role the pathways have in regulating urothelial phenotype, and investigate whether the pathways are implicated in muscle-invasive bladder cancer (MIBC). These aims were assessed using a cell culture paradigm of Normal Human Urothelial (NHU) cells that can be manipulated in vitro to represent different differentiated phenotypes, alongside MIBC cell lines and The Cancer Genome Atlas resource. Transcriptomic analysis of NHU cells identified a significant induction of VGLL1, a poorly understood regulator of Hippo signalling, in differentiated cells. Activation of upstream transcription factors PPARγ and GATA3 and/or blockade of active EGFR/RAS/RAF/MEK/ERK signalling were identified as mechanisms which induce VGLL1 expression in NHU cells. Ectopic overexpression of VGLL1 in undifferentiated NHU cells and MIBC cell line T24 resulted in significantly reduced proliferation. Conversely, knockdown of VGLL1 in differentiated NHU cells significantly reduced barrier tightness in an unwounded state, while inhibiting regeneration and increasing cell cycle activation in scratch-wounded cultures. A signalling pathway previously observed to be inhibited by VGLL1 function, YAP/TAZ, was unaffected by VGLL1 manipulation. -

Vitamin D Receptor Binding, Chromatin States and Association with Multiple Sclerosis
Human Molecular Genetics, 2012, Vol. 21, No. 16 3575–3586 doi:10.1093/hmg/dds189 Advance Access published on May 16, 2012 Vitamin D receptor binding, chromatin states and association with multiple sclerosis Giulio Disanto1,2,{, Geir Kjetil Sandve3,{, Antonio J. Berlanga-Taylor1,4, Giammario Ragnedda1,2,5, Julia M. Morahan1,2, Corey T. Watson1,2,6, Gavin Giovannoni7, George C. Ebers1,2 and Sreeram V. Ramagopalan1,2,7,8,∗ 1Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford, UK, 2Nuffield Department of Clinical Neurosciences (Clinical Neurology), University of Oxford, The West Wing, John Radcliffe Hospital, Oxford, UK, 3Department of Informatics, University of Oslo, Blindern, Oslo, Norway, 4Nuffield Department of Clinical Medicine, University of Oxford, John Radcliffe Hospital, Oxford, UK, 5Department of Clinical and Experimental Medicine, University of Sassari, Italy, 6Department of Biological Sciences, Simon Fraser University, Burnaby, BC, USA, 7Barts and The London School of Medicine and Dentistry, Blizard Institute, Queen Mary University of London, London, UK and 8London School of Hygiene and Tropical Medicine, London, UK Received April 23, 2012; Revised May 10, 2012; Accepted June 11, 2012 Both genetic and environmental factors contribute to the aetiology of multiple sclerosis (MS). More than 50 genomic regions have been associated with MS susceptibility and vitamin D status also influences the risk of this complex disease. However, how these factors interact in disease causation is unclear. We aimed to investigate the relationship between vitamin D receptor (VDR) binding in lymphoblastoid cell lines (LCLs), chromatin states in LCLs and MS-associated genomic regions. Using the Genomic Hyperbrowser, we found that VDR-binding regions overlapped with active regulatory regions [active promoter (AP) and strong enhancer (SE)] in LCLs more than expected by chance [45.3-fold enrichment for SE (P < 2.0e205) and 63.41-fold enrichment for AP (P < 2.0e205)]. -

Vertebrate Fatty Acid and Retinoid Binding Protein Genes and Proteins: Evidence for Ancient and Recent Gene Duplication Events
In: Advances in Genetics Research. Volume 11 ISBN: 978-1-62948-744-1 Editor: Kevin V. Urbano © 2014 Nova Science Publishers, Inc. Chapter 7 Vertebrate Fatty Acid and Retinoid Binding Protein Genes and Proteins: Evidence for Ancient and Recent Gene Duplication Events Roger S. Holmes Eskitis Institute for Drug Discovery and School of Biomolecular and Physical Sciences, Griffith University, Nathan, QLD, Australia Abstract Fatty acid binding proteins (FABP) and retinoid binding proteins (RBP) are members of a family of small, highly conserved cytoplasmic proteins that function in binding and facilitating the cellular uptake of fatty acids, retinoids and other hydrophobic compounds. Several human FABP-like genes are expressed in the body: FABP1 (liver); FABP2 (intestine); FABP3 (heart and skeletal muscle); FABP4 (adipocyte); FABP5 (epidermis); FABP6 (ileum); FABP7 (brain); FABP8 (nervous system); FABP9 (testis); and FABP12 (retina and testis). A related gene (FABP10) is expressed in lower vertebrate liver and other tissues. Four RBP genes are expressed in human tissues: RBP1 (many tissues); RBP2 (small intestine epithelium); RBP5 (kidney and liver); and RBP7 (kidney and heart). Comparative FABP and RBP amino acid sequences and structures and gene locations were examined using data from several vertebrate genome projects. Sequence alignments, key amino acid residues and conserved predicted secondary and tertiary structures were also studied, including lipid binding regions. Vertebrate FABP- and RBP- like genes usually contained 4 coding exons in conserved locations, supporting a common evolutionary origin for these genes. Phylogenetic analyses examined the relationships and evolutionary origins of these genes, suggesting division into three FABP gene classes: 1: FABP1, FABP6 and FABP10; 2: FABP2; and 3, with 2 groups: 3A: FABP4, FABP8, FABP9 and FABP12; and 3B: and FABP3, FABP5 and FABP7. -

Motifs in Saccharomyces Cerevisiae Master's Thesis
UNIVERSITY OF TARTU Faculty of Biology and Geography Institute of Molecular and Cell Biology Hedi Peterson The discovery and characterization of regulatory motifs in Saccharomyces cerevisiae Master's Thesis Supervisor: Jaak Vilo, PhD Tartu 2006 "Computers are useless. They can only give you answers." Pablo Picasso Contents 1 Introduction 1 1.1 Background . 1 1.2 Objective . 3 2 Background 4 2.1 Biological background . 4 2.1.1 Gene Ontology . 5 2.1.2 Regulatory motifs . 8 2.1.3 Protein-protein interactions . 8 2.2 Bioinformatics approaches . 10 2.2.1 Pattern Discovery . 10 2.2.2 Phylogenetic footprinting . 10 3 Material and methods 12 3.1 Data . 12 3.1.1 Sequences . 13 3.1.2 Previously known transcription factors and regulatory motifs . 15 3.1.3 GO groups . 16 3.1.4 Protein-protein interaction data . 17 3.2 Pattern discovery . 18 3.3 Statistical evaluation . 19 3.4 Expansion of groups by PPI data . 20 4 Results 23 4.1 Randomization . 23 4.2 Pattern discovery for known transcription factors' target groups 26 4.3 Pattern discovery in GO dataset . 27 4.4 Expansion of GO groups by PPI . 31 4.5 From protein-protein interactions to regulatory motifs and GO annotations . 31 4.5.1 Pipeline example . 32 4.6 Web-tool Gviz-PPI . 35 4.7 Data update to BiGeR . 36 5 Discussion 40 6 Conclusions 43 Summary 44 Summary in Estonian 46 Acknowledgements 48 References 49 Appendix 55 List of Figures 2.1 Example of a GO molecular function subtree. Here GO:0003887 is a leaf and GO:0003674 is a root node. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Variants at IRF5-TNPO3, 17Q12-21 and MMEL1 Are Associated with Primary Biliary Cirrhosis
University of Texas Rio Grande Valley ScholarWorks @ UTRGV Health and Biomedical Sciences Faculty Publications and Presentations College of Health Professions 8-2010 Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis Gideon M. Hirschfield Xiangdong Liu Younghun Han Ivan P. Gorlov Yan Lu See next page for additional authors Follow this and additional works at: https://scholarworks.utrgv.edu/hbs_fac Part of the Medicine and Health Sciences Commons Recommended Citation Hirschfield, G. M., Liu, X., Han, .,Y Gorlov, I. P., Lu, Y., Xu, C., Lu, Y., Chen, W., Juran, B. D., Coltescu, C., Mason, A. L., Milkiewicz, P., Myers, R. P., Odin, J. A., Luketic, V. A., Speiciene, D., Vincent, C., Levy, C., Gregersen, P. K., Zhang, J., … Siminovitch, K. A. (2010). Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nature genetics, 42(8), 655–657. https://doi.org/10.1038/ng.631 This Article is brought to you for free and open access by the College of Health Professions at ScholarWorks @ UTRGV. It has been accepted for inclusion in Health and Biomedical Sciences Faculty Publications and Presentations by an authorized administrator of ScholarWorks @ UTRGV. For more information, please contact [email protected], [email protected]. Authors Gideon M. Hirschfield, Xiangdong Liu, ounghunY Han, Ivan P. Gorlov, Yan Lu, Chun Xu, Yue Lu, Wei Chen, Brian D. Juran, and Catalina Coltescu This article is available at ScholarWorks @ UTRGV: https://scholarworks.utrgv.edu/hbs_fac/92 NIH Public Access Author Manuscript Nat Genet. Author manuscript; available in PMC 2010 August 27. -
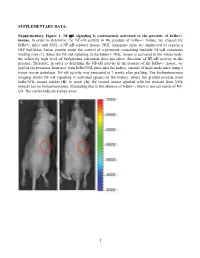
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age.