The Genetic Theory of Adaptation: a Brief History
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Natural Selection and Adaptation, Part I
Natural Selection and Adaptation, Part I 36-149 The Tree of Life Christopher R. Genovese Department of Statistics 132H Baker Hall x8-7836 http://www.stat.cmu.edu/~genovese/ . Plan • Review of Natural Selection • Detecting Natural Selection (discussion) • Examples of Observed Natural Selection ............................... Next Time: • Adaptive Traits • Methods for Reasoning about and studying adaptations • Explaining Complex Adaptations (discussion) 36-149 The Tree of Life Class #10 -1- Overview The theories of common descent and natural selection play different roles within the theory of evolution. Common Descent explains the unity of life. Natural Selection explains the diversity of life. An adaptation (or adaptive trait) is a feature of an organism that enhances reproductive success, relative to other possible variants, in a given environment. Adaptations become prevalent and are maintained in a population through natural selection. Indeed, natural selection is the only mechanism of evolutionary change that can satisfactorily explain adaptations. 36-149 The Tree of Life Class #10 -2- Darwin's Argument Darwin put forward two main arguments in support of natural selection: An analogical argument: Artificial selection A logical argument: The struggle for existence (As we will see later, we now have more than just argument in support of the theory.) 36-149 The Tree of Life Class #10 -3- The Analogical Argument: Artificial Selection 36-149 The Tree of Life Class #10 -4- The Analogical Argument: Artificial Selection 36-149 The Tree of Life Class #10 -5- The Analogical Argument: Artificial Selection Teosinte to Corn 36-149 The Tree of Life Class #10 -6- The Analogical Argument: Artificial Selection • Darwin was intimately familiar with the efforts of breeders in his day to produce novel varieties. -

1. Adaptation and the Evolution of Physiological Characters
Bennett, A. F. 1997. Adaptation and the evolution of physiological characters, pp. 3-16. In: Handbook of Physiology, Sect. 13: Comparative Physiology. W. H. Dantzler, ed. Oxford Univ. Press, New York. 1. Adaptation and the evolution of physiological characters Department of Ecology and Evolutionary Biology, University of California, ALBERT F. BENNETT 1 Irvine, California among the biological sciences (for example, behavioral CHAPTER CONTENTS science [I241). The Many meanings of "Adaptationn In general, comparative physiologists have been Criticisms of Adaptive Interpretations much more successful in, and have devoted much more Alternatives to Adaptive Explanations energy to, pursuing the former rather than the latter Historical inheritance goal (37). Most of this Handbook is devoted to an Developmentai pattern and constraint Physical and biomechanical correlation examination of mechanism-how various physiologi- Phenotypic size correlation cal systems function in various animals. Such compara- Genetic correlations tive studies are usually interpreted within a specific Chance fixation evolutionary context, that of adaptation. That is, or- Studying the Evolution of Physiological Characters ganisms are asserted to be designed in the ways they Macroevolutionary studies Microevolutionary studies are and to function in the ways they do because of Incorporating an Evolutionary Perspective into Physiological Studies natural selection which results in evolutionary change. The principal textbooks in the field (for example, refs. 33, 52, 102, 115) make explicit reference in their titles to the importance of adaptation to comparative COMPARATIVE PHYSIOLOGISTS HAVE TWO GOALS. The physiology, as did the last comparative section of this first is to explain mechanism, the study of how organ- Handbook (32). Adaptive evolutionary explanations isms are built functionally, "how animals work" (113). -

Lineage-Specific Mapping of Quantitative Trait Loci
Heredity (2013) 111, 106–113 & 2013 Macmillan Publishers Limited All rights reserved 0018-067X/13 www.nature.com/hdy ORIGINAL ARTICLE Lineage-specific mapping of quantitative trait loci C Chen1 and K Ritland We present an approach for quantitative trait locus (QTL) mapping, termed as ‘lineage-specific QTL mapping’, for inferring allelic changes of QTL evolution along with branches in a phylogeny. We describe and analyze the simplest case: by adding a third taxon into the normal procedure of QTL mapping between pairs of taxa, such inferences can be made along lineages to a presumed common ancestor. Although comparisons of QTL maps among species can identify homology of QTLs by apparent co- location, lineage-specific mapping of QTL can classify homology into (1) orthology (shared origin of QTL) versus (2) paralogy (independent origin of QTL within resolution of map distance). In this light, we present a graphical method that identifies six modes of QTL evolution in a three taxon comparison. We then apply our model to map lineage-specific QTLs for inbreeding among three taxa of yellow monkey-flower: Mimulus guttatus and two inbreeders M. platycalyx and M. micranthus, but critically assuming outcrossing was the ancestral state. The two most common modes of homology across traits were orthologous (shared ancestry of mutation for QTL alleles). The outbreeder M. guttatus had the fewest lineage-specific QTL, in accordance with the presumed ancestry of outbreeding. Extensions of lineage-specific QTL mapping to other types of data and crosses, and to inference of ancestral QTL state, are discussed. Heredity (2013) 111, 106–113; doi:10.1038/hdy.2013.24; published online 24 April 2013 Keywords: Quantitative trait locus mapping; evolution of mating systems; Mimulus INTRODUCTION common ancestor of these two taxa, back to the common ancestor of Knowledge of the genetic architecture of complex traits can offer all three taxa and forwards again to the third taxon. -
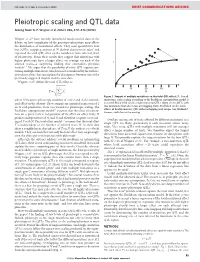
Pleiotropic Scaling and QTL Data Arising From: G
NATURE | Vol 456 | 4 December 2008 BRIEF COMMUNICATIONS ARISING Pleiotropic scaling and QTL data Arising from: G. P. Wagner et al. Nature 452, 470–473 (2008) Wagner et al.1 have recently introduced much-needed data to the debate on how complexity of the genotype–phenotype map affects the distribution of mutational effects. They used quantitative trait loci (QTLs) mapping analysis of 70 skeletal characters in mice2 and regressed the total QTL effect on the number of traits affected (level of pleiotropy). From their results they suggest that mutations with higher pleiotropy have a larger effect, on average, on each of the T affected traits—a surprising finding that contradicts previous models3–7. We argue that the possibility of some QTL regions con- taining multiple mutations, which was not considered by the authors, introduces a bias that can explain the discrepancy between one of the previously suggested models and the new data. Wagner et al.1 define the total QTL effect as vffiffiffiffiffiffiffiffiffiffiffiffiffi u uXN 10 20 30 ~t 2 T Ai N i~1 Figure 1 | Impact of multiple mutations on the total QTL effect, T. For all where N measures pleiotropy (number of traits) and Ai the normal- mutations, strict scaling according to the Euclidean superposition model is ized effect on the ith trait. They compare an empirical regression of T assumed. Black filled circles, single-mutation QTLs. Open circles, QTLs with on N with predictions from two models for pleiotropic scaling. The two mutations that affect non-overlapping traits. Red filled circles, total Euclidean superposition model3,4 assumes that the effect of a muta- effects of double-mutant QTL with overlapping trait ranges (see Methods). -

Plant Evolution an Introduction to the History of Life
Plant Evolution An Introduction to the History of Life KARL J. NIKLAS The University of Chicago Press Chicago and London CONTENTS Preface vii Introduction 1 1 Origins and Early Events 29 2 The Invasion of Land and Air 93 3 Population Genetics, Adaptation, and Evolution 153 4 Development and Evolution 217 5 Speciation and Microevolution 271 6 Macroevolution 325 7 The Evolution of Multicellularity 377 8 Biophysics and Evolution 431 9 Ecology and Evolution 483 Glossary 537 Index 547 v Introduction The unpredictable and the predetermined unfold together to make everything the way it is. It’s how nature creates itself, on every scale, the snowflake and the snowstorm. — TOM STOPPARD, Arcadia, Act 1, Scene 4 (1993) Much has been written about evolution from the perspective of the history and biology of animals, but significantly less has been writ- ten about the evolutionary biology of plants. Zoocentricism in the biological literature is understandable to some extent because we are after all animals and not plants and because our self- interest is not entirely egotistical, since no biologist can deny the fact that animals have played significant and important roles as the actors on the stage of evolution come and go. The nearly romantic fascination with di- nosaurs and what caused their extinction is understandable, even though we should be equally fascinated with the monarchs of the Carboniferous, the tree lycopods and calamites, and with what caused their extinction (fig. 0.1). Yet, it must be understood that plants are as fascinating as animals, and that they are just as important to the study of biology in general and to understanding evolutionary theory in particular. -

Evolutionary and Historical Biogeography of Animal Diversity Learning Objectives
Evolutionary and historical biogeography of animal diversity Learning objectives • The students can explain the common ancestor of animal kingdom. • The students can explain the historical biogeography of animal. • The students can explain the invasion of animal from aquatic to terrestrial habitat. • The students can explain the basic mechanism of speciation, allopatric and non-allopatric. The Common Ancestor of Animal Kingdom Characteristics of Animals • Animals or “metazoans” are typically heterotrophic, multicellular organisms with diploid, eukaryotic cells. • Trichoplax adhaerens is defined as an animal by the presence of different somatic (i.e., non-reproductive) cell types and by impermeable cell–cell connections. Trichoplax adhaerens Blackstone, 2009 Two Hypotheses for the Branching Order of Groups at the Root of the Metazoan Tree 1 2 The choanoflagellates serve as an outgroup in the Bilaterians are the sister group to the placozoan + analysis, and sponges are the sister group to the sponge + ctenophore + cnidarian clade, while placozoans placozoan + cnidarian + ctenophore + bilaterian are the sister group to the sponge + ctenophore + clade. cnidarian clade. Blackstone, 2009 Ancestry and evolution of animal–bacterial interactions • Choanoflagellates as the last common ancestor of animal kingdom. • Urmetazoan is the group of animal with multicellular and produce differentiated cell types (ex. Egg & sperm) R.A. Alegado & N. King, 2014 Conserved morphology and ultrastructure of Choanoflagellates and Sponge choanocytes The collar complex is conserved in choanoflagellates (A. S. rosetta) and sponge collar cells (B. Sycon coactum) flagellum (fL), microvilli (mv), a nucleus (nu), and a food vacuole (fv) Brunet & King, 2017 The Historical Biogeography of Animal Zoogeographic regions Old New Cox, 2001 Plate tectonic regulation of global marine animal diversity A. -

Evolution by Natural Selection, Formulated Independently by Charles Darwin and Alfred Russel Wallace
UNIT 4 EVOLUTIONARY PATT EVOLUTIONARY E RNS AND PROC E SS E Evolution by Natural S 22 Selection Natural selection In this chapter you will learn that explains how Evolution is one of the most populations become important ideas in modern biology well suited to their environments over time. The shape and by reviewing by asking by applying coloration of leafy sea The rise of What is the evidence for evolution? Evolution in action: dragons (a fish closely evolutionary thought two case studies related to seahorses) 22.1 22.4 are heritable traits that with regard to help them to hide from predators. The pattern of evolution: The process of species have changed evolution by natural and are related 22.2 selection 22.3 keeping in mind Common myths about natural selection and adaptation 22.5 his chapter is about one of the great ideas in science: the theory of evolution by natural selection, formulated independently by Charles Darwin and Alfred Russel Wallace. The theory explains how T populations—individuals of the same species that live in the same area at the same time—have come to be adapted to environments ranging from arctic tundra to tropical wet forest. It revealed one of the five key attributes of life: Populations of organisms evolve. In other words, the heritable characteris- This chapter is part of the tics of populations change over time (Chapter 1). Big Picture. See how on Evolution by natural selection is one of the best supported and most important theories in the history pages 516–517. of scientific research. -

Comparative Evolution: Latent Potentials for Anagenetic Advance (Adaptive Shifts/Constraints/Anagenesis) G
Proc. Natl. Acad. Sci. USA Vol. 85, pp. 5141-5145, July 1988 Evolution Comparative evolution: Latent potentials for anagenetic advance (adaptive shifts/constraints/anagenesis) G. LEDYARD STEBBINS* AND DANIEL L. HARTLtt *Department of Genetics, University of California, Davis, CA 95616; and tDepartment of Genetics, Washington University School of Medicine, Box 8031, 660 South Euclid Avenue, Saint Louis, MO 63110 Contributed by G. Ledyard Stebbins, April 4, 1988 ABSTRACT One of the principles that has emerged from genetic variation available for evolutionary changes (2), a experimental evolutionary studies of microorganisms is that major concern of modem evolutionists is explaining how the polymorphic alleles or new mutations can sometimes possess a vast amount of genetic variation that actually exists can be latent potential to respond to selection in different environ- maintained. Given the fact that in complex higher organisms ments, although the alleles may be functionally equivalent or most new mutations with visible effects on phenotype are disfavored under typical conditions. We suggest that such deleterious, many biologists, particularly Kimura (3), have responses to selection in microorganisms serve as experimental sought to solve the problem by proposing that much genetic models of evolutionary advances that occur over much longer variation is selectively neutral or nearly so, at least at the periods of time in higher organisms. We propose as a general molecular level. Amidst a background of what may be largely evolutionary principle that anagenic advances often come from neutral or nearly neutral genetic variation, adaptive evolution capitalizing on preexisting latent selection potentials in the nevertheless occurs. While much of natural selection at the presence of novel ecological opportunity. -

Response to Selection in Finite Locus Models with Nonadditive Effects
Journal of Heredity, 2017, 318–327 doi:10.1093/jhered/esw123 Original Article Advance Access publication January 12, 2017 Original Article Response to Selection in Finite Locus Models with Nonadditive Effects Hadi Esfandyari, Mark Henryon, Peer Berg, Jørn Rind Thomasen, Downloaded from https://academic.oup.com/jhered/article/108/3/318/2895120 by guest on 23 September 2021 Piter Bijma, and Anders Christian Sørensen From the Center for Quantitative Genetics and Genomics, Department of Molecular Biology and Genetics, Aarhus University DK-8830 Tjele, Denmark (Esfandyari, Berg, Thomasen, and Sørensen); Seges, Danish Pig Research Centre, Copenhagen, Denmark (Henryon); School of Animal Biology, University of Western Australia, Crawley, Australia (Henryon); Nordic Genetic Resource Center, Ås, Norway (Berg); VikingGenetics, Assentoft, Denmark (Thomasen); and Animal Breeding and Genomics Centre, Wageningen University, Wageningen, The Netherlands (Bijma). Address correspondence to H. Esfandyari at the address above, or e-mail: hadi.esfandyari@ mbg.au.dk. Received May 18, 2016; First decision June 7, 2016; Accepted January 2, 2017. Corresponding Editor: C Scott Baker Abstract Under the finite-locus model in the absence of mutation, the additive genetic variation is expected to decrease when directional selection is acting on a population, according to quantitative-genetic theory. However, some theoretical studies of selection suggest that the level of additive variance can be sustained or even increased when nonadditive genetic effects are present. We tested the hypothesis that finite-locus models with both additive and nonadditive genetic effects maintain more additive genetic variance (VA) and realize larger medium- to long-term genetic gains than models with only additive effects when the trait under selection is subject to truncation selection. -

Microevolution and the Genetics of Populations Microevolution Refers to Varieties Within a Given Type
Chapter 8: Evolution Lesson 8.3: Microevolution and the Genetics of Populations Microevolution refers to varieties within a given type. Change happens within a group, but the descendant is clearly of the same type as the ancestor. This might better be called variation, or adaptation, but the changes are "horizontal" in effect, not "vertical." Such changes might be accomplished by "natural selection," in which a trait within the present variety is selected as the best for a given set of conditions, or accomplished by "artificial selection," such as when dog breeders produce a new breed of dog. Lesson Objectives ● Distinguish what is microevolution and how it affects changes in populations. ● Define gene pool, and explain how to calculate allele frequencies. ● State the Hardy-Weinberg theorem ● Identify the five forces of evolution. Vocabulary ● adaptive radiation ● gene pool ● migration ● allele frequency ● genetic drift ● mutation ● artificial selection ● Hardy-Weinberg theorem ● natural selection ● directional selection ● macroevolution ● population genetics ● disruptive selection ● microevolution ● stabilizing selection ● gene flow Introduction Darwin knew that heritable variations are needed for evolution to occur. However, he knew nothing about Mendel’s laws of genetics. Mendel’s laws were rediscovered in the early 1900s. Only then could scientists fully understand the process of evolution. Microevolution is how individual traits within a population change over time. In order for a population to change, some things must be assumed to be true. In other words, there must be some sort of process happening that causes microevolution. The five ways alleles within a population change over time are natural selection, migration (gene flow), mating, mutations, or genetic drift. -

Directional Selection and Variation in Finite Populations
Copyright 0 1987 by the Genetics Society of America Directional Selection and Variation in Finite Populations Peter D. Keightley and ‘William G. Hill Institute of Animal Genetics, University of Edinburgh, West Mains Road, Edinburgh EH9 3JN, Scotland Manuscript received April 12, 1987 Revised copy accepted July 16, 1987 ABSTRACT Predictions are made of the equilibrium genetic variances and responses in a metric trait under the joint effects of directional selection, mutation and linkage in a finite population. The “infinitesimal model“ is analyzed as the limiting case of many mutants of very small effect, otherwise Monte Carlo simulation is used. If the effects of mutant genes on the trait are symmetrically distributed and they are unlinked, the variance of mutant effects is not an important parameter. If the distribution is skewed, unless effects or the population size is small, the proportion of mutants that have increasing effect is the critical parameter. With linkage the distribution of genotypic values in the population becomes skewed downward and the equilibrium genetic variance and response are smaller as disequilibrium becomes important. Linkage effects are greater when the mutational variance is contributed by many genes of small effect than few of large effect, and are greater when the majority of mutants increase rather than decrease the trait because genes that are of large effect or are deleterious do not segregate for long. The most likely conditions for ‘‘Muller’s ratchet” are investi- gated. N recent years there has been much interest in the is attained more quickly in the presence of selection I production and maintenance of variation in pop- than in its absence, and is highly dependent on popu- ulations by mutation, stimulated by the presence of lation size. -

Evolution, Third Edition
evolution THIRD EDITION DOUGLAS J. FUTUYMA Stony Brook University Chapter 20, "Evolution of Genes and Genomes" by Scott V. Edwards, Harvard University Chapter 21, "Evolution and Development" by John R. True, Stony Brook University SINAUER ASSOCIATES, INC. •Publishers Sunderland, Massachusetts U.S.A. © 2013 Sinauer Associates, Inc. This material cannot be copied, reproduced, manufactured or disseminated in any form without express written permission from the publisher. 00_EVOL3E_Frontmatter_U&lc.indd iii 2/14/13 8:05 AM Brief Contents 1 Evolutionary Biology 1 2 The Tree of Life: Classifi cation and Phylogeny 19 3 Patterns of Evolution 51 4 Evolution in the Fossil Record 77 5 A History of Life on Earth 103 6 The Geography of Evolution 135 7 The Evolution of Biodiversity 161 8 The Origin of Genetic Variation 189 9 Variation: The Foundation of Evolution 217 10 Genetic Drift: Evolution at Random 257 11 Natural Selection and Adaptation 281 12 The Genetic Theory of Natural Selection 309 13 Phenotypic Evolution 347 14 The Evolution of Life Histories 379 15 Sex and Reproductive Success 399 16 Confl ict and Cooperation 427 17 Species 459 18 Speciation 483 19 The Evolution of Interactions among Species 513 20 Evolution of Genes and Genomes 537 21 Evolution and Development 565 22 Macroevolution: Evolution above the Species Level 605 23 Evolutionary Science, Creationism, and Society 631 © 2013 Sinauer Associates, Inc. This material cannot be copied, reproduced, manufactured or disseminated in any form without express written permission from the publisher.