DNA Barcoding for the Identification of Smoked Fish Products
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TESIS DE DOCTORADO Desarrollo De Herramientas Moleculares Para Su Aplicación En La Mejora De La Trazabilidad De Los Alimentos Fátima C
TESIS DE DOCTORADO Desarrollo de herramientas moleculares para su aplicación en la mejora de la trazabilidad de los alimentos Fátima C. Lago Soriano 2017 Desarrollo de herramientas moleculares para para moleculares Desarrollo de herramientas : DO Fátima Soriano Lago C. TESIS DOCTORA DE la los trazabilidad de alimentos aplicaciónla su mejora de en 2017 Escuela Internacional de Doctorado Fátima C. Lago Soriano TESIS DE DOCTORADO DESARROLLO DE HERRAMIENTAS MOLECULARES PARA SU APLICACIÓN EN LA MEJORA DE LA TRAZABILIDAD DE LOS ALIMENTOS Dirigida por los Doctores: Montserrat Espiñeira Fernández Juan Manuel Vieites Baptista de Sousa Página 1 de 153 AGRADECIMIENTOS Cuando una etapa llega a su fin, es cuando por fin puedes mirar a atrás, respirar profundamente, y acordarte de aquellos que te acompañaron. Del mismo modo, es difícil entender los agradecimientos de una tesis hasta que pones el punto y final. Es en este momento cuando se puede percibir la gratitud que sientes a todas las personas que han estado presentes durante esa etapa, ya bien sea codo a codo o simplemente trayéndote un café calentito en el momento preciso. Pero también es cierto que, entre toda esa gente que ha estado ahí, hay pocas caras que se dibujan clara e intensamente en mi cabeza. En primerísimo lugar, me gustaría dar las gracias de una manera muy especial a Montse por muchos, muchísimos motivos: por darme cariño y amistad desde el día en que nos conocimos; porque a lo largo de esta década hemos compartido muchísimos momentos alegres, acompañados de risas y carcajadas, pero también los más tristes de mi vida, inundados de lágrimas y angustia; por estar ahí para lo que sea, para todo, y tener siempre tendida su mano amiga; por escucharme una y otra vez, sin cansarse, y aconsejarme sabiamente; por confiar en mí y guiarme, no solo durante el desarrollo de esta tesis, sino también en mi formación y día a día; por su eterna paciencia;… y, sobre todo, por poner en mi vida al “morenocho”, ese pequeño loquito tímido que me comería a besos. -

Distributions and Habitats: Anguillidae
Distributions and Habitats: Anguillidae FAMILY Anguillidae Rafinesque, 1810 - freshwater eels GENUS Anguilla Schrank, 1798 - freshwater eels Species Anguilla anguilla (Linnaeus, 1758) - common eel Distribution: Western and eastern Atlantic, Baltic Sea, North Sea, White Sea, Mediterranean Sea, Black Sea, Sea of Marmara: European seas and adjacent watersheds, spawing and larval migration routes to and from the western Atlantic. Habitat: freshwater, brackish, marine. Species Anguilla australis Richardson, 1841 - shortfinned eel Distribution: Southwestern Pacific. Habitat: freshwater, brackish, marine. Species Anguilla bengalensis (Gray, 1831) - mottled eel Distribution: Indian Ocean; southeastern Africa; Nepal, India Pakistan and Bangladesh. Habitat: freshwater, brackish, marine. Species Anguilla bicolor McClelland, 1844 - shortfin eel Distribution: Western Indian Ocean, Africa and India: South African and East African watersheds and islands in Western Indian Ocean (Seychelles, Madagascar and Mascarenes) east to India and Sri Lanka and to Western Australia, north to China. Habitat: freshwater, brackish, marine. Species Anguilla borneensis Popta, 1924 - Indonesian longfinned eel Distribution: Bo River, eastern Borneo. Habitat: freshwater, brackish, marine Species Anguilla celebesensis Kaup, 1856 - Celebes longfin eel Distribution: Western Pacific: Philippines to central Indonesia. Habitat: freshwater, brackish, marine. Species Anguilla dieffenbachii Gray, 1842 - New Zealand longfin eel Distribution: New Zealand. Habitat: freshwater, brackish, -

New Zealand Fishes a Field Guide to Common Species Caught by Bottom, Midwater, and Surface Fishing Cover Photos: Top – Kingfish (Seriola Lalandi), Malcolm Francis
New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing Cover photos: Top – Kingfish (Seriola lalandi), Malcolm Francis. Top left – Snapper (Chrysophrys auratus), Malcolm Francis. Centre – Catch of hoki (Macruronus novaezelandiae), Neil Bagley (NIWA). Bottom left – Jack mackerel (Trachurus sp.), Malcolm Francis. Bottom – Orange roughy (Hoplostethus atlanticus), NIWA. New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing New Zealand Aquatic Environment and Biodiversity Report No: 208 Prepared for Fisheries New Zealand by P. J. McMillan M. P. Francis G. D. James L. J. Paul P. Marriott E. J. Mackay B. A. Wood D. W. Stevens L. H. Griggs S. J. Baird C. D. Roberts‡ A. L. Stewart‡ C. D. Struthers‡ J. E. Robbins NIWA, Private Bag 14901, Wellington 6241 ‡ Museum of New Zealand Te Papa Tongarewa, PO Box 467, Wellington, 6011Wellington ISSN 1176-9440 (print) ISSN 1179-6480 (online) ISBN 978-1-98-859425-5 (print) ISBN 978-1-98-859426-2 (online) 2019 Disclaimer While every effort was made to ensure the information in this publication is accurate, Fisheries New Zealand does not accept any responsibility or liability for error of fact, omission, interpretation or opinion that may be present, nor for the consequences of any decisions based on this information. Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries website at http://www.mpi.govt.nz/news-and-resources/publications/ A higher resolution (larger) PDF of this guide is also available by application to: [email protected] Citation: McMillan, P.J.; Francis, M.P.; James, G.D.; Paul, L.J.; Marriott, P.; Mackay, E.; Wood, B.A.; Stevens, D.W.; Griggs, L.H.; Baird, S.J.; Roberts, C.D.; Stewart, A.L.; Struthers, C.D.; Robbins, J.E. -
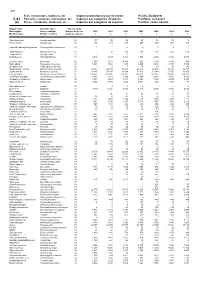
Fish, Crustaceans, Molluscs, Etc Capture Production by Species
478 Fish, crustaceans, molluscs, etc Capture production by species items Pacific, Southwest C-81 Poissons, crustacés, mollusques, etc Captures par catégories d'espèces Pacifique, sud-ouest (a) Peces, crustáceos, moluscos, etc Capturas por categorías de especies Pacífico, sudoccidental English name Scientific name Species group Nom anglais Nom scientifique Groupe d'espèces 2000 2001 2002 2003 2004 2005 2006 Nombre inglés Nombre científico Grupo de especies t t t t t t t Short-finned eel Anguilla australis 22 37 38 28 27 13 10 5 River eels nei Anguilla spp 22 380 313 337 267 209 277 210 Chinook(=Spring=King)salmon Oncorhynchus tshawytscha 23 1 1 0 4 1 2 1 Sand flounders Rhombosolea spp 31 ... 37 204 193 187 437 514 Tonguefishes Cynoglossidae 31 - - 3 - - - - Flatfishes nei Pleuronectiformes 31 2 954 3 234 2 818 3 308 2 980 3 766 3 050 Common mora Mora moro 32 1 358 1 211 1 308 1 234 1 403 1 154 986 Red codling Pseudophycis bachus 32 5 364 4 526 4 443 8 265 9 540 8 165 5 854 Grenadier cod Tripterophycis gilchristi 32 - 2 7 10 13 13 43 Southern blue whiting Micromesistius australis 32 43 419 49 982 72 203 43 812 26 576 30 304 32 735 Southern hake Merluccius australis 32 16 041 15 188 13 834 22 623 19 344 12 560 12 858 Blue grenadier Macruronus novaezelandiae 32 274 625 247 841 215 302 209 414 147 032 134 145 119 329 Thorntooth grenadier Lepidorhynchus denticulatus 32 3 833 4 783 5 349 5 304 6 341 3 855 4 056 Grenadiers, rattails nei Macrouridae 32 2 394 3 094 3 877 4 253 3 732 2 660 2 848 Gadiformes nei Gadiformes 32 2 853 5 479 3 252 3 281 298 1 217 47 Sea catfishes nei Ariidae 33 .. -

Checklist of Marine Demersal Fishes Captured by the Pair Trawl Fisheries in Southern (RJ-SC) Brazil
Biota Neotropica 19(1): e20170432, 2019 www.scielo.br/bn ISSN 1676-0611 (online edition) Inventory Checklist of marine demersal fishes captured by the pair trawl fisheries in Southern (RJ-SC) Brazil Matheus Marcos Rotundo1,2,3,4 , Evandro Severino-Rodrigues2, Walter Barrella4,5, Miguel Petrere Jun- ior3 & Milena Ramires4,5 1Universidade Santa Cecilia, Acervo Zoológico, R. Oswaldo Cruz, 266, CEP11045-907, Santos, SP, Brasil 2Instituto de Pesca, Programa de Pós-graduação em Aquicultura e Pesca, Santos, SP, Brasil 3Universidade Federal de São Carlos, Programa de Pós-Graduação em Planejamento e Uso de Recursos Renováveis, Rodovia João Leme dos Santos, Km 110, CEP 18052-780, Sorocaba, SP, Brasil 4Universidade Santa Cecília, Programa de Pós-Graduação de Auditoria Ambiental, R. Oswaldo Cruz, 266, CEP11045-907, Santos, SP, Brasil 5Universidade Santa Cecília, Programa de Pós-Graduação em Sustentabilidade de Ecossistemas Costeiros e Marinhos, R. Oswaldo Cruz, 266, CEP11045-907, Santos, SP, Brasil *Corresponding author: Matheus Marcos Rotundo: [email protected] ROTUNDO, M.M., SEVERINO-RODRIGUES, E., BARRELLA, W., PETRERE JUNIOR, M., RAMIRES, M. Checklist of marine demersal fishes captured by the pair trawl fisheries in Southern (RJ-SC) Brazil. Biota Neotropica. 19(1): e20170432. http://dx.doi.org/10.1590/1676-0611-BN-2017-0432 Abstract: Demersal fishery resources are abundant on continental shelves, on the tropical and subtropical coasts, making up a significant part of the marine environment. Marine demersal fishery resources are captured by various fishing methods, often unsustainably, which has led to the depletion of their stocks. In order to inventory the marine demersal ichthyofauna on the Southern Brazilian coast, as well as their conservation status and distribution, this study analyzed the composition and frequency of occurrence of fish captured by pair trawling in 117 fishery fleet landings based in the State of São Paulo between 2005 and 2012. -

Appendices Appendices
APPENDICES APPENDICES APPENDIX 1 – PUBLICATIONS SCIENTIFIC PAPERS Aidoo EN, Ute Mueller U, Hyndes GA, and Ryan Braccini M. 2015. Is a global quantitative KL. 2016. The effects of measurement uncertainty assessment of shark populations warranted? on spatial characterisation of recreational fishing Fisheries, 40: 492–501. catch rates. Fisheries Research 181: 1–13. Braccini M. 2016. Experts have different Andrews KR, Williams AJ, Fernandez-Silva I, perceptions of the management and conservation Newman SJ, Copus JM, Wakefield CB, Randall JE, status of sharks. Annals of Marine Biology and and Bowen BW. 2016. Phylogeny of deepwater Research 3: 1012. snappers (Genus Etelis) reveals a cryptic species pair in the Indo-Pacific and Pleistocene invasion of Braccini M, Aires-da-Silva A, and Taylor I. 2016. the Atlantic. Molecular Phylogenetics and Incorporating movement in the modelling of shark Evolution 100: 361-371. and ray population dynamics: approaches and management implications. Reviews in Fish Biology Bellchambers LM, Gaughan D, Wise B, Jackson G, and Fisheries 26: 13–24. and Fletcher WJ. 2016. Adopting Marine Stewardship Council certification of Western Caputi N, de Lestang S, Reid C, Hesp A, and How J. Australian fisheries at a jurisdictional level: the 2015. Maximum economic yield of the western benefits and challenges. Fisheries Research 183: rock lobster fishery of Western Australia after 609-616. moving from effort to quota control. Marine Policy, 51: 452-464. Bellchambers LM, Fisher EA, Harry AV, and Travaille KL. 2016. Identifying potential risks for Charles A, Westlund L, Bartley DM, Fletcher WJ, Marine Stewardship Council assessment and Garcia S, Govan H, and Sanders J. -

Table of Fishes of Sydney Harbour 2019
Table of Fishes of Sydney Harbour 2019 Family Family/Com Species Species Common Notes mon Name Name Acanthuridae Surgeonfishe Acanthurus Eyestripe close s dussumieri Surgeonfish to southern li mit Acanthuridae Acanthurus Orangebloch close to olivaceus Surgeonfish southern limit Acanthuridae Acanthurus Convict close to triostegus Surgeonfish southern limit Acanthuridae Acanthurus Yellowmask xanthopterus Surgeonfish Acanthuridae Paracanthurus Blue Tang not included hepatus in species count Acanthuridae Prionurus Spotted Sawtail maculatus Acanthuridae Prionurus Australian Sawtail microlepidotus Ambassidae Glassfishes Ambassis Port Jackson jacksoniensis glassfish Ambassidae Ambassis marianus Estuary Glassfish Anguillidae Freshwater Anguilla australis Shortfin Eel Eels Anguillidae Anguilla reinhardtii Longfinned Eel Antennariidae Anglerfishes Antennarius Freckled Anglerfish southern limit coccineus Antennariidae Antennarius Giant Anglerfish close to commerson southen limit Antennariidae Antennarius Shaggy Anglerfish southern limit hispidus Antennariidae Antennarius pictus Painted Anglerfish Antennariidae Antennarius striatus Striate Anglerfish Table of Fishes of Sydney Harbour 2019 Antennariidae Histrio histrio Sargassum close to Anglerfish southen limit Antennariidae Porophryne Red-fingered erythrodactylus Anglerfish Aploactinidae Velvetfishes Aploactisoma Southern Velvetfish milesii Aploactinidae Cocotropus Patchwork microps Velvetfish Aploactinidae Paraploactis Bearded Velvetfish trachyderma Aplodactylidae Seacarps Aplodactylus Rock Cale -

AUSTRALIAN NATIONAL SPEARFISHING RECORDS JULY 2018 COMPILED by AUF RECORDS OFFICER VIN RUSHWORTH Common Names
AUSTRALIAN NATIONAL SPEARFISHING RECORDS JULY 2018 COMPILED BY AUF RECORDS OFFICER VIN RUSHWORTH Common Names Common Name Pg. Common Name Pg. Albacore 37 Cod, Barramundi 39 Amberjack 8 Cod, Bearded 27 Amberjack, High-fin 9 Cod, Black 40 Angelfish Yellow-Mask 32 Cod, Blacksaddle Rock 40 Angelfish, Blue 32 Cod, Blackspotted 40 Angelfish, Imperial 32 Cod, Black-Tipped Rock 40 Angelfish, Six Banded 32 Cod, Break-Sea 39 Anglerfish, Spinycoat 3 Cod, Camouflage 40 Barracouta 13 Cod, Chinaman 41 Barracuda, Blackfin 44 Cod, Coral 39 Barracuda, Blue and Gold 44 Cod, Coral Rock 39 Barracuda, Chevron 44 Cod, Dusky 42 Barracuda, Great 44 Cod, Flowery 40 Barracuda, Pickhandle 44 Cod, Freckled Coral 39 Barramundi 20 Cod, Gold Spotted 39 Bass, Red 23 Cod, Highfin 40 Batfish, Black-tip 13 Cod, Long-finned Rock 41 Batfish, Hump-headed 13 Cod, Long-headed 41 Batfish, Long-finned 13 Cod, Maori 41 Batfish, Long-snout 13 Cod, Masked 41 Batfish, Short-finned 13 Cod, Ocellated 39 Bigeye, Lunar-tailed 33 Cod, Peacock Coral 39 Blackfish, Banded Rock 14 Cod, Potato 41 Blackfish, Rock 14 Cod, Pug-Nosed Wire-netting 40 Blackfish, Western Rock 14 Cod, Purple 40 Blanquillo, Blue 25 Cod, Rankin 40 Blue Devil, Eastern 31 Cod, Red 27 Blue Devil, Southern 31 Cod, Red Rock 37 Bluefish 13 Cod, Red Rock Western 38 Blue-lined Seabream 24 Cod, Red-flushed Rock 38 Boarfish, Giant 30 Cod, Speckledfin 40 Boarfish, Longsnout 30 Cod, Tomato Coral 39 Boarfish, Short 30 Cod, Twin-Spot 39 Boarfish, Yellow-spotted 30 Cod, White-lined Rock 38 Bonefish, Eastern 3 Coral Fish, New-moon -

Fish and Invertebrate Bycatch and Discards in New Zealand Hoki, Hake, and Ling Fisheries from 1990–91 Until 2012–13
Fish and invertebrate bycatch and discards in New Zealand hoki, hake, or ling trawl fisheries from 1990–91 until 2012–13 New Zealand Aquatic Environment and Biodiversity Report No. 163 S.L. Ballara R.L. O’Driscoll ISSN 1179-6480 (online) ISBN 978-1-77665-111-5 (online) November 2015 Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries websites at: http://www.mpi.govt.nz/news-resources/publications.aspx http://fs.fish.govt.nz go to Document library/Research reports © Crown Copyright - Ministry for Primary Industries Table of Contents EXECUTIVE SUMMARY 1 1. INTRODUCTION 3 2. METHODS 5 2.1 Definition of terms 5 2.2 Observer data 5 2.2.1 Data preparation and grooming 6 2.3 Commercial fishing return data 8 2.4 Analysis of factors influencing bycatch and discards 9 2.5 Calculation of bycatch and discard rates 9 2.6 Analysis of temporal trends in bycatch and discards 10 2.7 Comparison of trends in bycatch with data from trawl surveys 11 2.8 Discard information from Catch Landing Returns 11 2.9 Observer-authorised discarding 12 3. RESULTS 12 3.1 Distribution and representativeness of observer data 12 3.2 Comparison of estimators 13 3.3 Bycatch data (excluding discards) 14 3.3.1 Overview of raw bycatch data 14 3.3.2 Regression modelling and stratification of bycatch data 15 3.4 Discard data 15 3.4.1 Overview of -

A New Congrid Eel (Teleostei: Anguilliformes: Congridae) from the Western Pacific, with an Analysis of Its Relationships
Zootaxa 4845 (2): 191–210 ISSN 1175-5326 (print edition) https://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2020 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4845.2.2 http://zoobank.org/urn:lsid:zoobank.org:pub:B2DA6D79-874E-48C1-B054-FEC08546223C A new congrid eel (Teleostei: Anguilliformes: Congridae) from the Western Pacific, with an analysis of its relationships DAVID G. SMITH1*, EMMA S. KARMOVSKAYA2 & JOÃO PAULO CAPRETZ BATISTA DA SILVA3 1Smithsonian Institution, Museum Support Center, MRC-534, 4210 Silver Hill Road, Suitland, MD 20746 [email protected]; https://orcid.org/0000-0002-6354-2427 2Shirshov Institute of Oceanology, Russian Academy of Sciences, Moscow, 117218, Russia [email protected]; https://orcid.org/0000-0002-0636-4265 3Departamento de Sistemática e Ecologia, Centro de Ciências Exatas e da Natureza, Universidade Federal da Paraíba, Castelo Branco, 58051-900, João Pessoa, PB, Brazil [email protected]; https://orcid.org/0000-0002-2373-3421 *Corresponding author Abstract A new species of congrid eel, Bathycongrus villosus sp. nov., is described from the Philippines and Vanuatu. It is similar to some of the small-toothed species currently placed in Bathycongrus and to the species of Bassanago. In this paper we compare the new species to Bassanago albescens (Barnard, 1923) and to Bathycongrus parviporus Karmovskaya, 2011, which it most closely resembles. An analysis of 19 characters shows that it agrees with Bat. parviporus in 16 characters and with Bas. albescens in one. In two characters, the three species are all different. We therefore place it in Bathycongrus. Key words: Taxonomy, Pisces, Bathycongrus, new species Introduction The species described here was discovered independently by two of the authors. -

Reproductive Biology of Southwestern Atlantic Wreckfish, Polyprion
Environmental Biology of Fishes 68: 163–173, 2003. © 2003 Kluwer Academic Publishers. Printed in the Netherlands. Reproductive biology of southwestern Atlantic wreckfish, Polyprion americanus (Teleostei: Polyprionidae) Monicaˆ B. Peresa & Sandro Klippelb aServic¸o da Regiao˜ Litoral, DQA, FEPAM Rua Carlos Chagas 55 sala 707, Centro Porto Alegre, RS 90.030-020, Brazil (e-mail: [email protected]) bTalha-mar Projetos Ambientais, Cons. d’Avila 190 Porto Alegre, RS 91040-450, Brazil Received 5 February 2003 Accepted 12 July 2003 Key words: sexual maturity, fecundity, spawning aggregation, spawning season, migration, deep-sea fish, life-cycle closure, southern Brazil Synopsis We report on the reproductive biology of southwestern Atlantic wreckfish. Females mature first at 77.9 cm total length (TL) (10.4 years) and all are mature by 90 cm TL (15.2 years). Males mature first at 74.9 cm (9 years) and all are mature by 80 cm TL (10.9 years). The wreckfish is a gonochoristic multiple spawner and the gonadal cycle is synchronized at the population level. Spawning occurs from late July to early October along the continental slope (>300 m). Ovarian fecundity varies from 3 to 11.9 million (135–311 oocytes × g−1) and increases exponentially with length. Spawning at western boundary current systems, maintained by homing of adults, is a basic requirement for self-sustaining populations of this species. Introduction are mature and juveniles may live more than 2 years at the sea surface before recruitment to the bottom The wreckfish, Polyprion americanus (Bloch and (Sedberry et al. 1999). Despite its worldwide distri- Schneider, 1801), is a large, long-lived demersal teleost bution and high commercial importance, there is no that inhabits continental and oceanic island slopes of information on reproductive cycle, fecundity, spawn- temperate and subtropical waters of the northern and ing sites and season of these fish. -

Kaikōura Marine Area If It Specified in the Regulations Has Not Had One-Third of the Telson Cut Off
KAIKŌURA Changes to the MARINE recreational AREA fishing rules Sperm whale © Whale Watch Kaikoura Sperm whale © Whale Watch To help keep fish stocks sustainable and protect against illegal fishing and poaching, the Kaikōura Marine Area has been introduced. Within the Marine Area (blue line on the map) there are Marine changes to the recreational Area fishing rules. What are the new rules? » The recreational daily bag limits for some finfish and shellfish species have been reduced. » The size limits for blue cod and sea perch have changed. » Daily limits for the harvest of Sanctuary bladder kelp and karengo have Marine Reserve been introduced. Mātaitai Taiāpure » A prohibition on the taking of red Marine Area moki. » The introduction of ‘telson clipping’ and an accumulation limit for spiny rock lobster. Turn over for a summary of the new rules. These are not the full rules – for a full version visit www.mpi.govt.nz Finfish Maximum daily limit Minimum length Species What is the ‘telson clipping’ rule? per fisher (cm) The telson is the central part of the tail fan on a Blue cod 6 33 rock lobster. Tarakihi 10 25 Telson clipping is a way of marking spiny rock Sea Perch 20 26 lobster to make it clear that they have been Kahawai 10 No limit recreationally caught. One-third of the telson is cut Butterfish 10 35 off so that it is noticeably shorter than the other Red Moki NO TAKE sections of the tail fan. (See the picture below). Blue Moki 10 40 Rig 3 No limit School shark 3 No limit Kingfish 75 Bass and Hapuku Daily combined bag No limit limit of five per person Bluenose No limit with a maximum of 3 Ling of any one species No limit Albacore tuna No limit Game sharks (seven-gilled shark, mako shark, blue shark, 1 game shark No limit hammerhead shark, porbeagle total One-third of the shark, thresher shark) central telson Shellfish has been clipped.