Expression Analyses of Flower Developmental Genes in Eschscholzia Californica
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
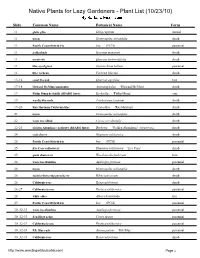
Native Plants for Lazy Gardeners - Plant List (10/23/10)
Native Plants for Lazy Gardeners - Plant List (10/23/10) Slide Common Name Botanical Name Form 11 globe gilia Gilia capitata annual 11 toyon Heteromeles arbutifolia shrub 11 Pacific Coast Hybrid iris Iris (PCH) perennial 11 goldenbush Isocoma menziesii shrub 11 scrub oak Quercus berberidifolia shrub 11 blue-eyed grass Sisyrinchium bellum perennial 11 lilac verbena Verbena lilacina shrub 13-16 coast live oak Quercus agrifolia tree 17-18 Howard McMinn man anita Arctostaphylos 'Howard McMinn' shrub 19 Philip Mun keckiella (RSABG Intro) Keckiella 'Philip Munz' ine 19 woolly bluecurls Trichostema lanatum shrub 19-20 Ray Hartman California lilac Ceanothus 'Ray Hartman' shrub 21 toyon Heteromeles arbutifolia shrub 22 western redbud Cercis occidentalis shrub 22-23 Golden Abundance barberry (RSABG Intro) Berberis 'Golden Abundance' (MAHONIA) shrub 2, coffeeberry Rhamnus californica shrub 25 Pacific Coast Hybrid iris Iris (PCH) perennial 25 Eve Case coffeeberry Rhamnus californica '. e Case' shrub 25 giant chain fern Woodwardia fimbriata fern 26 western columbine Aquilegia formosa perennial 26 toyon Heteromeles arbutifolia shrub 26 fuchsia-flowering gooseberry Ribes speciosum shrub 26 California rose Rosa californica shrub 26-27 California fescue Festuca californica perennial 28 white alder Alnus rhombifolia tree 29 Pacific Coast Hybrid iris Iris (PCH) perennial 30 032-33 western columbine Aquilegia formosa perennial 30 032-33 San Diego sedge Carex spissa perennial 30 032-33 California fescue Festuca californica perennial 30 032-33 Elk Blue rush Juncus patens '.l1 2lue' perennial 30 032-33 California rose Rosa californica shrub http://www weedingwildsuburbia com/ Page 1 30 032-3, toyon Heteromeles arbutifolia shrub 30 032-3, fuchsia-flowering gooseberry Ribes speciosum shrub 30 032-3, Claremont pink-flowering currant (RSA Intro) Ribes sanguineum ar. -

Epimedium L) – a Promising Source of Raw Materials for the Creation of Medicines for the Treatment of Erectile Dysfunction in Men
Pharmacogn J. 2020; 12(6)Suppl:1710-1715 A Multifaceted Journal in the field of Natural Products and Pharmacognosy Research Article www.phcogj.com Representatives of the Genus Goryanka (Epimedium L) – a Promising Source of Raw Materials for the Creation of Medicines for the Treatment of Erectile Dysfunction in Men Bukinich Darya Dmitrievna, Salova VG, Odintsova EB, Rastopchina OV, Solovyovа NL, Kozlova AM, Krasniuk II (jun), Krasniuk II, Kozlova Zh M* ABSTRACT Erectile dysfunction and multiple mechanisms of its development are one of the most pressing problems of modern medicine. In the twenty-first century, millions of men around the world suffer from sexual disorders, and the number of such patients is only growing from year to Bukinich Darya Dmitrievna, year. The flavonoid icariin, contained in plants of the genusEpimedium L., is a promising Salova VG, Odintsova EB, pharmacologically active substance used for erectile dysfunction, due to its ability to affect Rastopchina OV, Solovyovа type 5 phosphodiesterase, inhibiting its activity. To date, domestic and foreign pharmaceutical NL, Kozlova AM, Krasniuk II companies produce biologically active food additives and herbal preparations, which include (jun), Krasniuk II, Kozlova Zh Goryanka extract. But the range of standardized herbal medicines is very small. M* Key words: Drug, Epimedium Estrellita, Icariin, Impotence. First Moscow state medical university named after I.M. Sechenov, (Sechenov University), Moscow, RUSSIAN INTRODUCTION The purpose of this work is to theoretically FEDERATION. substantiate the possibility of using medicinal plant Erectile dysfunction (ED) is one of the most raw materials of the genus Goryanka (Epimedium Correspondence pressing problems of modern medicine. According L) to create medicines for the treatment of erectile Kozlova Zh. -

Survey of Roadside Alien Plants in Hawai`I Volcanoes National Park and Adjacent Residential Areas 2001–2005
Technical Report HCSU-032 SURVEY OF ROADSIDE ALIEN PLANts IN HAWAI`I VOLCANOES NATIONAL PARK AND ADJACENT RESIDENTIAL AREAS 2001–2005 Linda W. Pratt1 Keali`i F. Bio2 James D. Jacobi1 1 U.S. Geological Survey, Pacific Island Ecosystems Research Center, Kilauea Field Station, P.O. Box 44, Hawaii National Park, HI 96718 2 Hawai‘i Cooperative Studies Unit, University of Hawai‘i at Hilo, P.O. Box 44, Hawai‘i National Park, HI 96718 Hawai‘i Cooperative Studies Unit University of Hawai‘i at Hilo 200 W. Kawili St. Hilo, HI 96720 (808) 933-0706 September 2012 This product was prepared under Cooperative Agreement CA03WRAG0036 for the Pacific Island Ecosystems Research Center of the U.S. Geological Survey. Technical Report HCSU-032 SURVEY OF ROADSIDE ALIEN PLANTS IN HAWAI`I VOLCANOES NATIONAL PARK AND ADJACENT RESIDENTIAL AREAS 2001–2005 1 2 1 LINDA W. PRATT , KEALI`I F. BIO , AND JAMES D. JACOBI 1 U.S. Geological Survey, Pacific Island Ecosystems Research Center, Kīlauea Field Station, P.O. Box 44, Hawai`i Volcanoes National Park, HI 96718 2 Hawaii Cooperative Studies Unit, University of Hawai`i at Hilo, Hilo, HI 96720 Hawai`i Cooperative Studies Unit University of Hawai`i at Hilo 200 W. Kawili St. Hilo, HI 96720 (808) 933-0706 September 2012 This article has been peer reviewed and approved for publication consistent with USGS Fundamental Science Practices ( http://pubs.usgs.gov/circ/1367/ ). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. -

December 2015 / January 2016
University of Arizona Yavapai County Cooperative Extension Yavapai Gardens Master Gardener Newsletter December 2015 - January 2016 Mushrooms for Your Kitchen and Garden By Lori Dekker The world of mushrooms Events & Activities is entering a new era. In the past gardeners and MG Association Meeting, NO MEETING IN DECEM- foodies have considered BER, next meeting Jan. 20 in Prescott. 6:30pm the mushroom to be a Alta Vista Gardening Club, Prescott, fourth Tues- garden novelty or a tasty day of the month, 12:30pm. Call 928-458-9508 for culinary delight, while a information. few of us have an interest in the possible health benefits or the psy- cho/spiritual/recreational uses of a few of the more famous mush- Prescott Area Gourd Society, third Wednesday of the rooms. For now, I’d like to consider the potential health benefits of month, 10:30am, at Miller Valley Indoor Art Market, fungi in your soil and therefore your garden. 531 Madison Ave, Prescott When left to their own devices mushrooms, or more accu- rately fungi, are decomposers and eventually constructors. In a nut- Prescott Orchid Society, 4rd Sunday of the month, 1pm at the Prescott Library, (928) 717-0623 shell, they build soil from the raw material of litter and waste found in the garden. Since they digest food outside their bodies, they are Prescott Area Iris Society call 928-445-8132 for date essentially “sweating” digestive enzymes and producing waste as and place information. they grow through their environment. To put it more simply and hap- pily for gardeners, the fungus breaks down complex compounds Mountain View Garden Club, Prescott Valley, Dewey into more simple ones that then become available, leaving behind area, 2nd Friday of month, 1:30pm, call 775-4993 for metabolites that can, in turn, be utilized by other microbes. -

Perennial 2018 Report
2018 Perennial Trial Evaluation Department of Horticulture and Landscape Architecture, Colorado State University Scientific Name Acanthus longifolius Source Denver Botanic Gardens Common Name Bear's Breech Tsnum: 96147 Variety Bed: B Cultivar Family Acanthaceae Plant Type: Herbaceous perennial Plant Habit: Upright spreading clump Plant Quality: Good Foliage Color: Medium green Plant Height: 10 inches Plant Width: Around 4 feet Cultural Problems: Nuisances: Insect Problems: Disease Problems: Landscape Uses: Specimen, border Flower Effectiveness: Good Flower Color: Mauve and greenish white Flower Size: Inflorescence up to 10 inches in size Flower Height: Up to 4 feet Flower Effectiveness: Good Comments on plant: 3/8 5/18 budding 6/7 70 7/5 100 8/2 100 9/12 10/3 3/20 5/22 Hailstor 6/20 100 7/18 100 8/15 100 9/26 10/17 4/4 8/29 60 4/22 budding Page 1 of 1013 2018 Perennial Trial Evaluation Department of Horticulture and Landscape Architecture, Colorado State University Scientific Name Achillea Source Blooms of Bressingham Common Name Hybrid Yarrow Tsnum: 09141 Variety Bed: GB Cultivar 507 Family Asteraceae Plant Type: Herbaceous Plant Habit: Upright Plant Quality: Good Foliage Color: Medium Green Plant Height: 8-10 inches Plant Width: About 1 foot Cultural Problems: Nuisances: Insect Problems: Disease Problems: Landscape Uses: Border, mass, cut flowers fresh or dried Flower Effectiveness: Good Flower Color: Light Peach to Pink Flower Size: clusters up to 5 inches Flower Height: Up to 2 feet Flower Effectiveness: Good Comments on plant: 3/8 -

Landscape Plants Rated by Deer Resistance
E271 Bulletin For a comprehensive list of our publications visit www.rce.rutgers.edu Landscape Plants Rated by Deer Resistance Pedro Perdomo, Morris County Agricultural Agent Peter Nitzsche, Morris County Agricultural Agent David Drake, Ph.D., Extension Specialist in Wildlife Management The following is a list of landscape plants rated according to their resistance to deer damage. The list was compiled with input from nursery and landscape professionals, Cooperative Extension personnel, and Master Gardeners in Northern N.J. Realizing that no plant is deer proof, plants in the Rarely Damaged, and Seldom Rarely Damaged categories would be best for landscapes prone to deer damage. Plants Occasionally Severely Damaged and Frequently Severely Damaged are often preferred by deer and should only be planted with additional protection such as the use of fencing, repellents, etc. Success of any of these plants in the landscape will depend on local deer populations and weather conditions. Latin Name Common Name Latin Name Common Name ANNUALS Petroselinum crispum Parsley Salvia Salvia Rarely Damaged Tagetes patula French Marigold Ageratum houstonianum Ageratum Tropaeolum majus Nasturtium Antirrhinum majus Snapdragon Verbena x hybrida Verbena Brugmansia sp. (Datura) Angel’s Trumpet Zinnia sp. Zinnia Calendula sp. Pot Marigold Catharanthus rosea Annual Vinca Occasionally Severely Damaged Centaurea cineraria Dusty Miller Begonia semperflorens Wax Begonia Cleome sp. Spider Flower Coleus sp. Coleus Consolida ambigua Larkspur Cosmos sp. Cosmos Euphorbia marginata Snow-on-the-Mountain Dahlia sp. Dahlia Helichrysum Strawflower Gerbera jamesonii Gerbera Daisy Heliotropium arborescens Heliotrope Helianthus sp. Sunflower Lobularia maritima Sweet Alyssum Impatiens balsamina Balsam, Touch-Me-Not Matricaria sp. False Camomile Impatiens walleriana Impatiens Myosotis sylvatica Forget-Me-Not Ipomea sp. -

Thesis Draft Rough
Wesleyan University The Honors College Plant-pollinator interactions across California grassland and coastal scrub vegetation types on San Bruno Mountain, San Mateo County by Miles Gordon Brooks Class of 2020 A thesis submitted to the faculty of Wesleyan University in partial fulfillment of the requirements for the Degree of Bachelor of Arts with Departmental Honors from the College of the Environment Middletown, Connecticut April, 2020 1 2 Abstract Animal pollination of plants is a crucial ecosystem service for maintaining biodiversity and ecosystem function, worldwide. High pollinator abundance and diversity can likewise improve the reproductive success of the plant community. Plant-pollinator interaction networks have the potential to identify dominant, specialist, and generalist pollinator species within a system, and their host plant counterparts. Understanding these relationships is paramount for buffering natural systems from biodiversity loss in a world where pollinator abundance continues to decline rapidly. San Bruno Mountain (SBM) in San Mateo County, California, is one of the last natural, open spaces in the urban landscape in the northern San Francisco Peninsula. I conducted a series of timed meanders and vegetation surveys at eight sample sites within SBM (four grassland and four coastal scrub sites) to identify plant species prevalence and pollinator species visitation of flowering plants. I employed a multivariate approach for investigating plant and pollinator species richness, plant and pollinator community composition, and trophic-level interactions across the SBM landscape, and I evaluated differences in these relationships between grassland and coastal scrub habitats. A total of 59 pollinator species and 135 plant species were inventoried over the course of the study. -

Cool Season Annuals
Cool Season Annuals HORT 308/609 Assigned Readings for Plant List 6 Plant List 6 Spring 2020 Read the pages in your textbook associated with the family descriptions and individual taxa covered on Plant List 6 that was distributed in lab. These plant lists are also available on the course website All Text And Images Are Copyrighted By: Dr. Michael A. Arnold, Texas A&M University, Dept. Horticultural Sciences, College Station, TX 77843-2133 Cool season flowers Cool Season A bit of landscaping helps Annuals most any structure! • Tolerant of freezing to subfreezing temperatures – Suitable for use throughout winter in southern half of our region – Suitable for late fall and very early spring use in northern portions of the region • Provides off-season color in winter Cool season foliage Cool (Season) Thoughts Alcea rosea • Many species are derived from edible or Hollyhocks medicinal European species • Classic old-fashioned reseeding annual, • Plants utilized solely for foliage are biennial, or weak perennial more common than with other • Tolerates cold to USDA z. 5, but heat of z. 8 is tough seasonal annuals • Tall cool season annuals are infrequent, • Bold coarse textured foliage; rounded mound the or become tall only late in the season first year or winter and then stiffly upright in spring • Limited range of soil moisture is common • Most decline when day temperatures consistently exceed 80°F or night temperatures exceed 70°F • Mostly for detail designs, bedding, or seasonal containers • Miniature hibiscus-like flowers – Singles quaint, -

Creating a Unified Landscape for the Widener Visitor's Center
University of Pennsylvania ScholarlyCommons Internship Program Reports Education and Visitor Experience 5-2009 Creating a Unified Landscape for the Widener Visitor’s Center M. E. Speller Follow this and additional works at: https://repository.upenn.edu/morrisarboretum_internreports Recommended Citation Speller, M. E., "Creating a Unified Landscape for the Widener Visitor’s Center" (2009). Internship Program Reports. 97. https://repository.upenn.edu/morrisarboretum_internreports/97 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/morrisarboretum_internreports/97 For more information, please contact [email protected]. Creating a Unified Landscape for the Widener Visitor’s Center This report is available at ScholarlyCommons: https://repository.upenn.edu/morrisarboretum_internreports/97 Title: Creating a Unified Landscape for the Widener Visitor’s Center Author: M.E. Speller Horticulture Intern Date: March 22, 2009 Abstract: The Widener Visitor’s Center is our primary point of contact with the public, however the landscape in front of the building fails to highlight its importance. Although this building supports many crucial functions of the Arboretum, in part because it is a re-purposed carriage house, the landscape around it easily becomes confusing and disjointed. Creating a cohesive design for the front of the Visitor Center will help visitors navigate both the building and the arboretum itself with greater ease. In addition, this landscape can be an example of good horticulture at its simplest, serving as a model for home landscaping. The new plan for Widener is designed to orient the visitor toward the front entrance where Visitor Services can best help him or her. The plants used were chosen primarily for soil adaptability and compact size, as well as year-round interest. -

A Legacy of Plants N His Short Life, Douglas Created a Tremendous Legacy in the Plants That He Intro (P Coulteri) Pines
The American lIorHcullural Sociely inviles you Io Celehrate tbe American Gardener al our 1999 Annual Conference Roston" Massachusetts June 9 - June 12~ 1999 Celebrate Ute accompHsbenls of American gardeners in Ute hlsloric "Cay Upon lhe 1Iill." Join wah avid gardeners from. across Ute counlrg lo learn new ideas for gardening excellence. Attend informa-Hve ledures and demonslraHons by naHonally-known garden experts. Tour lhe greal public and privale gardens in and around Roslon, including Ute Arnold Arborelum and Garden in Ute Woods. Meet lhe winners of AIlS's 1999 naHonJ awards for excellence in horHcullure. @ tor more informaHon, call1he conference regislrar al (800) 777-7931 ext 10. co n t e n t s Volume 78, Number 1 • '.I " Commentary 4 Hellebores 22 Members' Forum 5 by C. Colston Burrell Staghorn fern) ethical plant collecting) orchids. These early-blooming pennnials are riding the crest of a wave ofpopularity) and hybridizers are News from AHS 7 busy working to meet the demand. Oklahoma Horticultural Society) Richard Lighty) Robert E. Lyons) Grecian foxglove. David Douglas 30 by Susan Davis Price Focus 9 Many familiar plants in cultivation today New plants for 1999. are improved selections of North American species Offshoots 14 found by this 19th-century Scottish expLorer. Waiting for spring in Vermont. Bold Plants 37 Gardeners Information Service 15 by Pam Baggett Houseplants) transplanting a ginkgo tree) Incorporating a few plants with height) imposing starting trees from seed) propagating grape vines. foliage) or striking blossoms can make a dramatic difference in any landscape design. Mail-Order Explorer 16 Heirloom flowers and vegetables. -

Chapter 3 — Basic Botany
Chapter 3 BASIC BOTANY IDAHO MASTER GARDENER UNIVERSITY OF IDAHO EXTENSION Introduction 2 Plant Nomenclature and Classification 2 Family 3 Genus 3 Species 3 Variety and Cultivar 3 Plant Life Cycles 6 Annuals 6 Biennials 6 Perennials 6 Plant Parts and Their Functions 7 Vegetative Parts: Leaves, Stems, and Roots 7 Reproductive Parts: Flowers, Fruits, and Seeds 10 Plant Development 14 Seed Germination 14 Vegetative Growth Stage 14 Reproductive Growth Stage 14 Senescence 15 Further Reading and Resources 15 CHAPTER 3 BASIC BOTANY 3 - 1 Chapter 3 Basic Botany Jennifer Jensen, Extension Educator, Boundary County Susan Bell, Extension Educator, Ada County William Bohl, Extension Educator, Bingham County Stephen Love, Consumer Horticulture Specialist, Aberdeen Research and Extension Center Illustrations by Jennifer Jensen INTRODUCTION varying from country to country, region to region, and sometimes even within a local area. This makes Botany is the study of plants. To become a it difficult to communicate about a plant. For knowledgeable plant person, it is essential to example, the state flower of Idaho is Philadelphus understand basic plant science. It is important to lewisii , commonly called syringa in Idaho. In other understand how plants grow, how their various parts parts of the country, however, the same plant is function, how they are identified and named, and known as mock orange. To add to the confusion, how they interact with their environment. Learning Syringa is the genus for lilac shrubs. Another the language of botany means learning many new example of confusing common names is Malva words. Making the effort to learn this material will parviflora , which is called little mallow, round leaf prove extremely valuable and will create excitement mallow, cheeseweed, or sometimes buttonweed. -

Floristic Composition, Life Form and Chorology of Plant Life at Al-Saoda, Asir Region, South-Western Saudi Arabia
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by International Institute for Science, Technology and Education (IISTE): E-Journals Journal of Biology, Agriculture and Healthcare www.iiste.org ISSN 2224-3208 (Paper) ISSN 2225-093X (Online) Vol.4, No.26, 2014 Floristic Composition, Life Form and Chorology of Plant Life at Al-Saoda, Asir Region, South-Western Saudi Arabia Saadiya S. Seraj 1, Rahma N. Jrais 2 and, Sakeena K. Ayyad. 1 1, Assistant Prof. of plant ecology, Faculty of Science, King Khalid university. 2, Lecturer of plant Taxonomy, Faculty of Science, King Khalid university. Email: [email protected] Abstract Asir highlands constitute a major part of south-western Saudi Arabia, and have a temperate climate at elevations above 2500 m a.s.l. This area has a complicated topography. The variations in elevation and topography have resulted in distinctive vegetational zones. Floristic composition, life form and chorology of plant life at Al- Saoda region south – western Saudi Arabia was studied. Three major wadis (sites) were investigated , every site was represented by three localities representing up-stream , mid-stream and down-stream portions of each site. Nine field trips were carried out during 18 months .Vegetation of three localities ( Wadi Tahlal , Wadi Al- Moght and Beni Mazen ) was studied . Results revealed that the region considers a hot-spot in the Kingdom in term of plant diversity and more diverse compared with other well studied regions in Saudi Arabia. A total of 103 plant species belonging to 40 families were recorded in study area.