Tracking and Forecasting Milepost Moments of the Epidemic in the Early-Outbreak: Framework and Applications to the COVID-19 [Version 2; Peer Review: 2 Approved]
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Osprey Nest Queen Size Page 2 LC Cutting Correction
Template Layout Sheets Organizing, Bags, Foundation Papers, and Template Layout Sheets Sort the Template Layout Sheets as shown in the graphics below. Unit A, Temp 1 Unit A, Temp 1 Layout UNIT A TEMPLATE LAYOUT SHEET CUT 3" STRIP BACKGROUND FABRIC E E E ID ID ID S S S Please read through your original instructions before beginning W W W E E E Sheet. Place (2) each in Bag S S S TEMP TEMP TEMP S S S E E A-1 A-1 A-1 E W W W S S S I I I D D D E E E C TEMP C TEMP TEMP U U T T A-1 A-1 T A-1 #6, #7, #8, and #9 L L I the queen expansion set. The instructions included herein only I C C C N N U U U T T T T E E L L L I I I N N N E E replace applicable information in the pattern. E Unit A, Temp 2, UNIT A TEMPLATE LAYOUT SHEET Unit A, Temp 2 Layout CUT 3" STRIP BACKGROUND FABRIC S S S E E E The Queen Foundation Set includes the following Foundation Papers: W W W S S S ID ID ID E TEMP E TEMP E TEMP Sheet. Place (2) each in Bag A-2 A-2 A-2 E E E D D D I I TEMP I TEMP TEMP S S S A-2 A-2 A-2 W W W C E C E E S S S U C C U C U U U T T T T T T L L L L IN IN L IN I I N #6, #7, #8, and #9 E E N E E NP 202 (Log Cabin Full Blocks) ~ 10 Pages E NP 220 (Log Cabin Half Block with Unit A Geese) ~ 2 Pages NP 203 (Log Cabin Half Block with Unit B Geese) ~ 1 Page Unit B, Temp 1, ABRIC F BACKGROUND Unit B, Temp 1 Layout E E T SHEE YOUT LA TE TEMPLA A T UNI E D I D I D I S S STRIP 3" T CU S W W E W TP 101 (Template Layout Sheets for Log Cabins and Geese) ~ 2 Pages E S S E S TEMP TEMP TEMP S S S E E E 1 A- 1 A- 1 A- W W W S S S I I I D D D E E E C C Sheet. -

Here You Can Taste Wuhan Featured Food
Contents Basic Mandarin Chinese Words and Phrases............................................... 2 Useful Sayings....................................................................................... 2 In Restaurants....................................................................................... 3 Numbers................................................................................................3 Dinning and cafes..........................................................................................5 Eating Out in Wuhan.............................................................................5 List of Restaurants and Food Streets (sort by distance).......................6 4 Places Where You Can Taste Wuhan Featured Food.........................7 Restaurants and cafes in Walking Distance........................................11 1 Basic Mandarin Chinese Words and Phrases Useful Sayings nǐ hǎo Hello 你 好 knee how zài jiàn Goodbye 再 见 zi gee’en xiè xiè Thank You 谢 谢! sheh sheh bú yòng le, xiè xiè No, thanks. 不 用 了,谢 谢 boo yong la, sheh sheh bú yòng xiè You are welcome. 不 用 谢 boo yong sheh wǒ jiào… My name is… 我 叫… wore jeow… shì Yes 是 shr bú shì No 不 是 boo shr hǎo Good 好 how bù hǎo Bad 不 好 boo how duì bù qǐ Excuse Me 对 不 起 dway boo chee wǒ tīng bù dǒng I do not understand 我 听 不 懂 wore ting boo dong duō shǎo qián How much? 多 少 钱? dor sheow chen Where is the xǐ shǒu jiān zài nǎ lǐ See-sow-jian zai na-lee washroom? 洗 手 间 在 哪 里 2 In Restaurants In China, many people call a male waiter as handsome guy and a female waitress as beautiful girl. It is also common to call “fú wù yuan” for waiters of both genders. cài dān Menu 菜 单 tsai dan shuài gē Waiter(Handsome) 帅 哥 shuai ge měi nǚ Waitress(Beautiful) 美 女 may nyu fú wù yuán Waiter/Waitress 服 务 员 fu woo yuan wǒ xiǎng yào zhè ge 我 I would like this. -
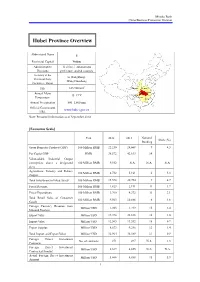
Hubei Province Overview
Mizuho Bank China Business Promotion Division Hubei Province Overview Abbreviated Name E Provincial Capital Wuhan Administrative 12 cities, 1 autonomous Divisions prefecture, and 64 counties Secretary of the Li Hongzhong; Provincial Party Wang Guosheng Committee; Mayor 2 Size 185,900 km Shaanxi Henan Annual Mean Hubei Anhui 15–17°C Chongqing Temperature Hunan Jiangxi Annual Precipitation 800–1,600 mm Official Government www.hubei.gov.cn URL Note: Personnel information as of September 2014 [Economic Scale] Unit 2012 2013 National Share (%) Ranking Gross Domestic Product (GDP) 100 Million RMB 22,250 24,668 9 4.3 Per Capita GDP RMB 38,572 42,613 14 - Value-added Industrial Output (enterprises above a designated 100 Million RMB 9,552 N.A. N.A. N.A. size) Agriculture, Forestry and Fishery 100 Million RMB 4,732 5,161 6 5.3 Output Total Investment in Fixed Assets 100 Million RMB 15,578 20,754 9 4.7 Fiscal Revenue 100 Million RMB 1,823 2,191 11 1.7 Fiscal Expenditure 100 Million RMB 3,760 4,372 11 3.1 Total Retail Sales of Consumer 100 Million RMB 9,563 10,886 6 4.6 Goods Foreign Currency Revenue from Million USD 1,203 1,219 15 2.4 Inbound Tourism Export Value Million USD 19,398 22,838 16 1.0 Import Value Million USD 12,565 13,552 18 0.7 Export Surplus Million USD 6,833 9,286 12 1.4 Total Import and Export Value Million USD 31,964 36,389 17 0.9 Foreign Direct Investment No. -

China Clean Energy Study Tour for Urban Infrastructure Development
China Clean Energy Study Tour for Urban Infrastructure Development BUSINESS ROUNDTABLE Tuesday, August 13, 2019 Hyatt Centric Fisherman’s Wharf Hotel • San Francisco, CA CONNECT WITH USTDA AGENDA China Urban Infrastructure Development Business Roundtable for U.S. Industry Hosted by the U.S. Trade and Development Agency (USTDA) Tuesday, August 13, 2019 ____________________________________________________________________ 9:30 - 10:00 a.m. Registration - Banquet AB 9:55 - 10:00 a.m. Administrative Remarks – KEA 10:00 - 10:10 a.m. Welcome and USTDA Overview by Ms. Alissa Lee - Country Manager for East Asia and the Indo-Pacific - USTDA 10:10 - 10:20 a.m. Comments by Mr. Douglas Wallace - Director, U.S. Department of Commerce Export Assistance Center, San Francisco 10:20 - 10:30 a.m. Introduction of U.S.-China Energy Cooperation Program (ECP) Ms. Lucinda Liu - Senior Program Manager, ECP Beijing 10:30 a.m. - 11:45 a.m. Delegate Presentations 10:30 - 10:45 a.m. Presentation by Professor ZHAO Gang - Director, Chinese Academy of Science and Technology for Development 10:45 - 11:00 a.m. Presentation by Mr. YAN Zhe - General Manager, Beijing Public Transport Tram Corporation 11:00 - 11:15 a.m. Presentation by Mr. LI Zhongwen - Head of Safety Department, Shenzhen Metro 11:15 - 11:30 a.m. Tea/Coffee Break 11:30 - 11:45 a.m. Presentation by Ms. WANG Jianxin - Deputy General Manager, Tianjin Metro Operation Corporation 11:45 a.m. - 12:00 p.m. Presentation by Mr. WANG Changyu - Director of General Engineer's Office, Wuhan Metro Group 12:00 - 12:15 p.m. -

High Speed Rail: Wuhan Urban Garden 5-Day Trip
High Speed Rail: Wuhan Urban Garden 5-Day Trip Day 1 Itinerary Suggested Transportation Hong Kong → Wuhan High Speed Rail [Hong Kong West Kowloon Station → Wuhan Railway Station] To hotel: Recommend to stay in a hotel by the river in Wuchang District. Metro: From Wuhan Railway Hotel for reference: Station, take Metro Line 4 The Westin Wuhan Wuchang Hotel towards Huangjinkou. Address: 96 Linjiang Boulevard, Wuchang District, Wuhan Change to Line 2 at Hongshan Square Station towards Tianhe International Airport. Get off at Jiyuqiao Station and walk for about 7 minutes. (Total travel time about 46 minutes) Taxi: About 35 minutes. Enjoy lunch near the hotel On foot: Walk for about 5 minutes Restaurant for reference: Zhen Bafang Hot Pot from the hotel. Address: No. 43 & 44, Building 12-13, Qianjin Road, Wanda Plaza, Jiyu Bridge, Wuchang District, Wuhan Stand the Test of Time: Yellow Crane Tower Bus: Walk for about 4 minutes from the restaurant to Jiyuqiao Metro Station. Take bus 804 towards Nanhu Road Jiangnan Village. Get off at Yue Ma Chang Station and walk for about 6 minutes. (Total travel time about 37 minutes) Taxi: About 15 minutes. Known as “The No. 1 Tower in the World”, the Yellow Crane Tower is a landmark for Wuhan City and Hubei Province and a must-see attraction. The tower was built in the Three Kingdoms era and was named after its erection on Huangjiji, a submerged rock. Well-known ancient characters such as Li Bai, Bai Juyi, Lu You and Yue Fei had all referenced the tower in their poetry works. -

Leading New ICT Building a Smart Urban Rail
Leading New ICT Building A Smart Urban Rail 2017 HUAWEI TECHNOLOGIES CO., LTD. Bantian, Longgang District Shenzhen518129, P. R. China Tel:+86-755-28780808 Huawei Digital Urban Rail Solution Digital Urban Rail Solution LTE-M Solution 04 Next-Generation DCS Solution 10 Urban Rail Cloud Solution 15 Huawei Digital Urban Rail Solution Huawei Digital Urban Rail Solution Huawei LTE-M Solution for Urban Rail Huawei and Alstom the Completed World’s Huawei Digital Urban Rail LTE-M Solution First CBTC over LTE Live Pilot On June 29th, 2015, Huawei and Alstom, one of the world’s leading energy solutions and transport companies, announced the successful completion of the world’s first live pilot test of 4G LTE multi-services based on Communications- based Train Control (CBTC), a railway signalling system based on wireless ground-to-train CBTC PIS CCTV Dispatching communication. The successful pilot, which CURRENT STATUS IN URBAN RAIL covered the unified multi-service capabilities TV Wall ATS Server Terminal In recent years, public Wi-Fi access points have become a OCC of several systems including CBTC, Passenger popular commodity in urban areas. Due to the explosive growth Information System (PIS), and closed-circuit in use of multimedia devices like smart phones, tablets and NMS LTE CN television (CCTV), marks a major step forward in notebooks, the demand on services of these devices in crowded the LTE commercialization of CBTC services. Line/Station Section/Depot Station places such as metro stations has dramatically increased. Huge BBU numbers of Wi-Fi devices on the platforms and in the trains RRU create chances of interference with Wi-Fi networks, which TAU TAU Alstom is the world’s first train manufacturer to integrate LTE 4G into its signalling system solution, the Urbalis Fluence CBTC Train AR IPC PIS AP TCMS solution, which greatly improves the suitability of eLTE, providing a converged ground-to-train wireless communication network Terminal When the CBTC system uses Wi-Fi technology to implement for metro operations. -

Table of Contents
International Conference on Soft Computing and Machine Learning (SCML2019) April 26th-29th, 2019, Wuhan, China Content Part I Conference Schedule ................................................................................................................................ 2 Part II Keynote Speeches .................................................................................................................................... 5 Keynote Speech 1: Fuzzy Logic and Signal Processing/Machine Learning for Brain-Computer Interface (BCI) ......................................................................................................................................................................... 5 Keynote Speech 2: Soft Computing and Machine Learning Services to Realize and Understand More About Our World ........................................................................................................................................................ 6 Part III Oral Sessions ........................................................................................................................................... 7 Oral Session_1 Deep Neural Networks ........................................................................................................... 8 Oral Session_2 Soft Computing ....................................................................................................................... 9 Oral Session_3 Machine Learning ................................................................................................................ -

Use Style: Paper Title
2019 4th International Conference on Education and Social Development (ICESD 2019) ISBN: 978-1-60595-621-3 Wuhan Subway Economic Development in the Perspective of "Innovation, Coordination, Green, Open, Sharing" Concept 1,a 2,b, Fang WANG and Xiao-sheng LEI * 1Hubei University of Traditional Chinese Medicine, Wuhan, Hubei, China 2Hubei University of Traditional Chinese Medicine, Wuhan, Hubei, China [email protected], [email protected] *Corresponding author Keywords: Development Concept, Wuhan, Subway Economy. Abstract. Since the 18th congress of the communist party of China, the "innovation, coordination, green, open and shared" development concept has been a profound change in the overall situation of China's development. In this context, the Wuhan Metro has developed rapidly, This paper analyzes the current situation, characteristics and influence of the development of subway economy in Wuhan from the perspective of development concept, and puts forward some suggestions for the future development of subway economy. Introduction For a long time, China has faced problems such as uneven, inadequate, uncoordinated and unsustainable development, and it has already faced the "ceiling dilemma". In recent years, China is facing the downward pressure of the economy. In order to change the development mode and solve the development problems, the 18th National Congress put forward five development concepts of “innovation, coordination, green, openness and sharing”, which have important guidance for the future development of social economy. Xiong'an New District is an innovative, coordinated, green ecological, open and co-constructed shared future urban model. Wuhan, following the glory of history, is becoming a reactivated city, and it must follow this development philosophy. -

Executive Management's Report: Overseas Growth
MTR CORPORATION LIMITED ANNUAL REPORT 2005 27 growingbroader The RMB4.6 billion Beijing Metro Line 4 and RMB6 billion Shenzhen Metro Line 4 projects, both major milestones on our overseas expansion, saw significant progress Overseas growth MTR Corporation made significant strides in our international expansion in 2005, achieving a number of major milestones in the Mainland of China. Mainland of China MTR Corporation’s entry into the rail market in the Mainland of China was heralded by the signing of an Agreement in Principle in January 2004 with the Shenzhen Municipal Government to build Phase 2 of the Shenzhen Metro Line 4 and to operate both Phase 1 and Phase 2 of the line for 30 years. With total project cost of Phase 2 estimated at RMB6 billion, the project also envisages significant property development at stations and depots along the new extension with an aggregate gross floor area of 2.9 million square metres of residential and commercial space based on the Company’s well proven “rail and property”business model in Hong Kong. During 2005, the Company achieved several major milestones on this build-operate-transfer project. In May, the feasibility study report was submitted to NDRC for approval, and the Concession Agreement was initialled with the Shenzhen Municipal Government, following which detailed design work began in earnest.This was followed in November by ground breaking on a trial section of Phase 2, with full project construction planned to commence once the Concession Agreement is signed. Bank financing for the project, representing approximately 60% of the total project cost, was secured in June when a detailed term sheet providing for a RMB 3.6 billion 25-year non-recourse bank loan was initialled with China Development Bank. -

Development of High-Speed Rail in the People's Republic of China
ADBI Working Paper Series DEVELOPMENT OF HIGH-SPEED RAIL IN THE PEOPLE’S REPUBLIC OF CHINA Pan Haixiao and Gao Ya No. 959 May 2019 Asian Development Bank Institute Pan Haixiao is a professor at the Department of Urban Planning of Tongji University. Gao Ya is a PhD candidate at the Department of Urban Planning of Tongji University. The views expressed in this paper are the views of the author and do not necessarily reflect the views or policies of ADBI, ADB, its Board of Directors, or the governments they represent. ADBI does not guarantee the accuracy of the data included in this paper and accepts no responsibility for any consequences of their use. Terminology used may not necessarily be consistent with ADB official terms. Working papers are subject to formal revision and correction before they are finalized and considered published. The Working Paper series is a continuation of the formerly named Discussion Paper series; the numbering of the papers continued without interruption or change. ADBI’s working papers reflect initial ideas on a topic and are posted online for discussion. Some working papers may develop into other forms of publication. Suggested citation: Haixiao, P. and G. Ya. 2019. Development of High-Speed Rail in the People’s Republic of China. ADBI Working Paper 959. Tokyo: Asian Development Bank Institute. Available: https://www.adb.org/publications/development-high-speed-rail-prc Please contact the authors for information about this paper. Email: [email protected] Asian Development Bank Institute Kasumigaseki Building, 8th Floor 3-2-5 Kasumigaseki, Chiyoda-ku Tokyo 100-6008, Japan Tel: +81-3-3593-5500 Fax: +81-3-3593-5571 URL: www.adbi.org E-mail: [email protected] © 2019 Asian Development Bank Institute ADBI Working Paper 959 Haixiao and Ya Abstract High-speed rail (HSR) construction is continuing at a rapid pace in the People’s Republic of China (PRC) to improve rail’s competitiveness in the passenger market and facilitate inter-city accessibility. -

Force Estimation Based on Semg Using Wavelet Analysis and Neural Network
Force Estimation Based on sEMG using Wavelet Analysis and Neural Network line 1: 1st Du Jiang line 1: 2ndGongfa Li line 1: 3rd Guozhang Jiang line 1: 4th Disi Chen line 2: Key Laboratory of line 2: Key Laboratory of line 2: Hubei Key Laboratory of line 2: School of Computing, Metallurgical Equipment and Metallurgical Equipment and Mechanical Transmission and University of Portsmouth Control Technology, Ministry of Control Technology, Ministry of Manufacturing Engineering, line3: Portsmouth, UK Education Wuhan University of Education Wuhan University of Wuhan University of Science and line 4: [email protected] Science and Technology Science and Technology Technology line 3: Hubei Key Laboratory of line 3: Research Center of line 3: 3D Printing and Mechanical Transmission and Biologic Manipulator and Intelligent Manufacturing line 1: 5th Zhaojie Ju Manufacturing Engineering, Intelligent Measurement and Engineering Institute, Wuhan line 2: School of Computing, Wuhan University of Science and Control, Wuhan University of University of Science and University of Portsmouth Technology Science and Technology Technology line 3: Portsmouth, UK line 4: Wuhan, China line 4: Wuhan, China line 4: Wuhan, China line 4: [email protected] line 5: [email protected] line 5: [email protected] line 5: [email protected] Abstract—In order to meet the needs of sEMG signal control gesture recognition and human-computer interaction[9]. The in human-computer interaction, an estimation of grip force based main work of sEMG-based human hand motion pattern on wavelet analysis and neural network is proposed. Firstly, the recognition is to study the action pattern of identifying the acquisition of EMG signals and the extraction methods of hand from the surface EMG signal, that is, the feature traditional features are described based on the introduction extraction and motion pattern recognition of sEMG[10], [11]. -

Travel Information
Travel Information Emergency Contact: Ms. MA Ya: +86 15527890078 Dr. MAO Yaxian: +86 15971483675 Prof. ZHOU Daicui: +86 15871350167 1. To Joyee Holiday Hotel(至卓悦酒店) Address: No.45 ZhuoDaoQuan Road, HongShan District, Wuhan,China / 武汉市洪山区卓刀泉路 45 号。 电话/Tel.:+86-27-87051888 Airport to Joyee Holiday Hotel (机场至卓悦酒店) 1) Taxi: Airport Highway → 2nd Yangtze River bridge or Er-qi Yangtze River bridge → ShuiGuoHu Tunnel → JieDaoKou Exit → LuoYu Road → ZhuoDaoQuanLiJiao → ZhuoDaoQuan Road → Joyee Holiday Hotel. It takes about 1 hour. The fare is about 150 RMB. 出租车:走机场高速,经长江二桥或二七长江大桥、水果湖隧道、街道口出口、 珞瑜路、卓刀泉立交桥、卓刀泉路、卓悦酒店。时间约1小时,车费150元左 右。 2) Subway: From terminal 3 take Subway Line 2 (Optics Valley direction), to HuQuan station (exit D). Walk about 5 minutes to the north. 地铁:可在3号航站楼乘坐地铁2号线到虎泉站(D出口),向北走5分钟即可到达 卓悦酒店。 Wuhan Railway Station to Joyee Holiday Hotel (武汉火车站至卓悦酒店) 1) Taxi: 2nd Ring Road → ShuiGuoHu Tunnel → JieDaoKou Exit → LuoYu Road → ZhuoDaoQuanLiJiao → ZhuoDaoQuan Road → Joyee Holiday Hotel. It takes about 30min. The fare is about 80 RMB. 出租车:走二环线、水果湖隧道、街道口出口、珞瑜路、卓刀泉立交桥、卓刀泉 路、卓悦酒店。30分钟车程,约80元车费。 2) Subway: take Subway Line 4 (HuangJinKou direction) transfer to line 2 at ZhongNanLu station (Optics Valley direction),to Huquan station (exit D). Walk about 5 minutes to the north. 地铁:乘地铁4号线(黄金口方向)到中南路站换乘地铁2号线到虎泉站(D出口),向 北走5分钟即可到达卓悦酒店。 Wuchang Railway Station to Joyee Holiday Hotel (武昌火车站至卓悦酒店) 1) Taxi: It takes about 20 min. The fare is about 25 RMB. 出租车:大约需要15分钟,花费25元。 2) Subway: take Subway Line 4 (Wuhan railway station direction) transfer to line 2 at ZhongNan road station (Optics Valley direction), to Huquan station (exit D). Walk about 5 minutes to the north.