Hyperspectral Data As a Biodiversity Screening Tool Can Differentiate
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hábitos Alimenticios De Pygocentrus Cariba Y Chalceus Epakros (Pisces, Characiformes: Characidae) En Dos Localidades De La Baja Orinoquia Colombiana
Memoria de la Fundación La Salle de Ciencias Naturales 2006 (“2005”), 164: 129-141 Hábitos alimenticios de Pygocentrus cariba y Chalceus epakros (Pisces, Characiformes: Characidae) en dos localidades de la baja Orinoquia colombiana Javier A. Maldonado-Ocampo y Hernando Ramírez-Gil Resumen. Este trabajo analiza la dieta de Pygocentrus cariba y Chalceus epakros (Characidae), especies capturadas entre mayo de 1998 y junio de 1999, en sistemas de la baja Orinoquia, Colombia. Para P. cariba se analizaron 156 individuos adultos, encontrándose siete categorías de alimento. A pesar de presentar un espectro variado de ítems alimenticios, unos pocos son dominantes, siendo los restos de peces (IIR= 38,38%; IAi= 91,1%) y los restos de material vegetal (IIR= 3,09%; IAi= 7,3%) los más ingeridos, aunque éste último puede ser consumido accidentalmente. En cuanto a la variación estacional en la dieta, los restos de peces fueron la categoría principal aunque no se encontraron diferencias estadísticamente significativas entre períodos en el índice de importancia relativa (IIR) o en el índice alimentario (IAi). Se analizaron 163 individuos de C. epakros que presentaron cuatro categorías de alimento, de las cuales dos, restos de material vegetal (IIR= 29,88%; IAi= 56,6%) e invertebrados (IIR= 22,72%; IAi= 43%) fueron las de mayor consumo en ambos períodos, no obstante, se presentaron diferencias estadísticamente diferentes entre épocas. Ambas especies presentan una dieta omnívora (oportunista), aunque P. cariba con tendencia piscívora y C. epakros con tendencia herbívora. Palabras clave. Ecología trófica. Arari. Caribe pechi rojo. Orinoco. Colombia. Feedings habits of Pygocentrus cariba and Chalceus epakros (Pisces, Characiformes: Characidae) in two localities from the lower Orinoquia in Colombia Abstract. -

§4-71-6.5 LIST of CONDITIONALLY APPROVED ANIMALS November
§4-71-6.5 LIST OF CONDITIONALLY APPROVED ANIMALS November 28, 2006 SCIENTIFIC NAME COMMON NAME INVERTEBRATES PHYLUM Annelida CLASS Oligochaeta ORDER Plesiopora FAMILY Tubificidae Tubifex (all species in genus) worm, tubifex PHYLUM Arthropoda CLASS Crustacea ORDER Anostraca FAMILY Artemiidae Artemia (all species in genus) shrimp, brine ORDER Cladocera FAMILY Daphnidae Daphnia (all species in genus) flea, water ORDER Decapoda FAMILY Atelecyclidae Erimacrus isenbeckii crab, horsehair FAMILY Cancridae Cancer antennarius crab, California rock Cancer anthonyi crab, yellowstone Cancer borealis crab, Jonah Cancer magister crab, dungeness Cancer productus crab, rock (red) FAMILY Geryonidae Geryon affinis crab, golden FAMILY Lithodidae Paralithodes camtschatica crab, Alaskan king FAMILY Majidae Chionocetes bairdi crab, snow Chionocetes opilio crab, snow 1 CONDITIONAL ANIMAL LIST §4-71-6.5 SCIENTIFIC NAME COMMON NAME Chionocetes tanneri crab, snow FAMILY Nephropidae Homarus (all species in genus) lobster, true FAMILY Palaemonidae Macrobrachium lar shrimp, freshwater Macrobrachium rosenbergi prawn, giant long-legged FAMILY Palinuridae Jasus (all species in genus) crayfish, saltwater; lobster Panulirus argus lobster, Atlantic spiny Panulirus longipes femoristriga crayfish, saltwater Panulirus pencillatus lobster, spiny FAMILY Portunidae Callinectes sapidus crab, blue Scylla serrata crab, Samoan; serrate, swimming FAMILY Raninidae Ranina ranina crab, spanner; red frog, Hawaiian CLASS Insecta ORDER Coleoptera FAMILY Tenebrionidae Tenebrio molitor mealworm, -

Los Peces Caribes De Venezuela.Pdf
Bol. Acad. C. Fís., Mat. y Nat. Vol. LXII No. 1 Marzo, 2002: 35-88. Antonio Machado-Allison: Los Peces Caribes de Venezuela LOS PECES CARIBES DE VENEZUELA: UNA APROXIMACIÓN A SU ESTUDIO TAXONÓMICO Antonio Machado Allison* El Trabajo presenta una sinopsis detallada de las diez y seis especies de caribes (pirañas) de Venezuela, incluidas en los géneros: Pygopristis (1 especie), Pristobrycon (4 especies), Pygocentrus (1 especie) y Serrasalmus (10 especies). Se discuten los aspectos histórico-taxonómico de cada especie, desde las Crónicas de Indias, los primeros naturalistas, hasta las contribuciones científicas más recientes. Se discute la validez de los nombres utilizados tradicionalmente y sus sinonimias. Se sugieren aspectos evolutivos y de relaciones filogenéticas intra y entre los diferentes géneros. Se ilustra cada especie con dibujos y fotografías y se incorporan claves para la identificación de los géneros y las especies. This paper present a detailed sinopsis of the piranha sixteen species of Venezuela, included in the genera: Pygopristis (1 species), Pristobrycon (4 species), Pygocentrus (1 species) y Serrasalmus (10 species). Discussions on historical-taxonomic aspects of each species are included from the Indian Crónicas de Indias, the first naturalists, to recent scientific contributions. Names traditionally used and sinonomies of each species are discussed. Suggestions on hypothesis of relationships and evolution inside groups and among genera are given. Each species is illustrated with drawings and photographs. Keys for the identification of species are included. Palabras Clave: Peces, Caribes, Venezuela, Clasificación Keywords: Fish, Piranha, Venezuela, Clasification I. INTRODUCCION mundial, gracias a la proliferación de historias, leyendas y fantasías muchas veces sin sentido. -
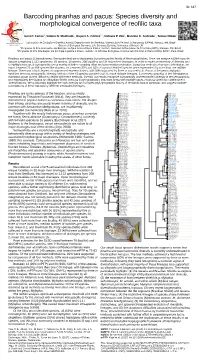
Barcoding Piranhas and Pacus: Species Diversity and Morphological Convergence of Reofilic Taxa
ID: 617 Barcoding piranhas and pacus: Species diversity and morphological convergence of reofilic taxa Izeni P. Farias1, Valeria N. Machado1, Rupert A. Collins1,2, Rafaela P. Ota3, Marcelo C. Andrade4, Tomas Hrbek1 1Laboratório de Evolução e Genética Animal, Departamento de Genética, Universidade Federal do Amazonas (UFAM), Manaus, AM, Brazil 2School of Biological Sciences, Life Sciences Building, University of Bristol, UK 3Programa de Pós-Graduação em Biologia de Água Doce e Pesca Interior, Instituto Nacional de Pesquisas da Amazônia (INPA), Manaus, AM, Brazil 4Programa de Pós-Graduação em Ecologia Aquática e Pesca, Instituto de Ciências Biológicas, Universidade Federal do Pará (UFPA), Belém, Pará, Brazil Piranhas and pacus (Characiformes: Serrasalmidae) are a charismatic but understudied family of Neotropical fishes. Here, we analyse a DNA barcode dataset comprising 1,122 specimens, 69 species, 16 genera, 208 localities and 34 major river drainages, in order to make an inventory of diversity and to highlight taxa and biogeographic areas worthy of further sampling effort and conservation protection. Using four methods of species delimitation, we report between 76 and 99 species-like clusters, i.e. between 20% and 33% of a priori identified species were represented by more than one mtDNA lineage. There was a high degree of congruence between clusters, with 60% supported by three or four methods. Pacus of the genus Myloplus exhibited the most intraspecific diversity, with six of the 13 species sampled found to have multiple lineages. Conversely, piranhas of the Serrasalmus rhombeus group proved difficult to delimit with these methods. Overall, our results recognize substantially underestimated diversity in the serrasalmids, and emphasizes the Guiana and Brazilian Shield rivers as biogeographically important areas with multiple cases of across-shield and within-shield diversifications. -

Universite Montpellier Ii Sciences Et Techniques Du Languedoc
UNIVERSITE MONTPELLIER II SCIENCES ET TECHNIQUES DU LANGUEDOC THESE Pour obtenir le grade de DOCTEUR DE L’UNIVERSITE DE MONTPELLIER II Discipline : Ecologie et Evolution Ecole Doctorale : SIBAGHE (Systèmes Intégrés en Biologie, Agronomie, Géosciences, Hydrosciences, Environnement). 2009 N°. Présentée et soutenue publiquement par TORRICO BALLIVIAN JUAN PABLO JOSÉ Le Titre Evolution de l’Ichtyofaune Amazonienne par l’approche de la phylogéographie comparée. Jury : M. BONHOMME François Directeur de Thèse M. DESMARAIS Erick Examinateur M. DOUZERY Emmanuel Examinateur M. LECOINTRE Guillaume Rapporteur M. MONTOYA-BURGOS Juan I. Rapporteur M. RENNO Jean-François Co-directeur de Thèse 2 A la mémoire de Jorge A. Torrico B. ( †10 janvier 2009) 3 A ma famille Jorge Torrico Q. Gilka Ballivian C. Cecilia Torrico B. Grace Morgan de Torrico Jorge Torrico Morgan Matias Dereck Torrico Morgan María Lucia Centellas 4 Cette thèse a été réalisée dans le cadre de programme IRD «Caractérisation et Valorisation de Ichtyofaune amazonienne pour une Aquaculture Raisonnée » en coopération avec l’Institut de Biologie Moleculaire (IBMB) de l’Université San Andrés à La Paz (Bolivie) et avec l’Institut des Sciences de l’Evolution de Montpellier (Isem). Elle a pu être possible grâce à la réalisation au préalable d’une maestria dans le laboratoire de l’IBMB et de deux stages préparatoires dans le laboratoire « Gènomes, Populations, Interactions, Adaptation » de l’ISEM (GPIA, 2001 et 2002). L’IRD a financé la majeure partie des recherches et des déplacements, l’ISEM une partie des recherches et l’hébergement sur sa station de biologie marine à Sète, l’ambassade de France en Bolivie a octroyé une demi-bourse. -
(Characiformes, Serrasalmidae) from the Rio Madeira Basin, Brazil
A peer-reviewed open-access journal ZooKeys 571: 153–167Myloplus (2016) zorroi, a new serrasalmid species from Madeira river basin... 153 doi: 10.3897/zookeys.571.5983 RESEARCH ARTICLE http://zookeys.pensoft.net Launched to accelerate biodiversity research A new large species of Myloplus (Characiformes, Serrasalmidae) from the Rio Madeira basin, Brazil Marcelo C. Andrade1,2, Michel Jégu3, Tommaso Giarrizzo1,2,4 1 Universidade Federal do Pará, Cidade Universitária Prof. José Silveira Netto. Laboratório de Biologia Pe- squeira e Manejo dos Recursos Aquáticos, Grupo de Ecologia Aquática. Avenida Perimetral, 2651, Terra Firme, 66077830. Belém, PA, Brazil 2 Programa de Pós-Graduação em Ecologia Aquática e Pesca. Universidade Fe- deral do Pará, Instituto de Ciências Biológicas. Cidade Universitária Prof. José Silveira Netto. Avenida Augusto Corrêa, 1, Guamá, 66075110. Belém, PA, Brazil 3 Institut de Recherche pour le Développement, Biologie des Organismes et Ecosystèmes Aquatiques, UMR BOREA, Laboratoire d´Icthyologie, Muséum national d’Histoire naturelle, MNHN, CP26, 43 rue Cuvier, 75231 Paris Cedex 05, France 4 Programa de Pós-Graduação em Biodiversidade e Conservação. Universidade Federal do Pará, Faculdade de Ciências Biológicas. Avenida Cel. José Porfírio, 2515, São Sebastião, 68372010. Altamira, PA, Brazil Corresponding author: Marcelo C. Andrade ([email protected]) Academic editor: C. Baldwin | Received 7 March 2015 | Accepted 19 January 2016 | Published 7 March 2016 http://zoobank.org/A5ABAD5A-7F31-46FB-A731-9A60A4AA9B83 Citation: Andrade MC, Jégu M, Giarrizzo T (2016) A new large species of Myloplus (Characiformes, Serrasalmidae) from the Rio Madeira basin, Brazil. ZooKeys 571: 153–167. doi: 10.3897/zookeys.571.5983 Abstract Myloplus zorroi sp. n. -

Dynamics of Ovarian Maturation During the Reproductive Cycle of Metynnis Maculatus, a Reservoir Invasive Fish Species (Teleostei: Characiformes)
Neotropical Ichthyology, 11(4):821-830, 2013 Copyright © 2013 Sociedade Brasileira de Ictiologia Dynamics of ovarian maturation during the reproductive cycle of Metynnis maculatus, a reservoir invasive fish species (Teleostei: Characiformes) Thiago Scremin Boscolo Pereira1, Renata Guimarães Moreira2 and Sergio Ricardo Batlouni1 In this study, we evaluated the dynamics of ovarian maturation and the spawning processes during the reproductive cycle of Metynnis maculatus. Adult females (n = 36) were collected bimonthly between April 2010 and March 2011. The mean gonadosomatic index (GSI) was determined, ovarian and blood samples were submitted for morphometric evaluation and the steroid plasma concentration was determined by ELISA. This species demonstrated asynchronous ovarian development with multiple spawns. This study revealed that, although defined as a multiple spawning species, the ovaries ofM. maculatus have a pattern of development with a predominance of vitellogenesis between April and August and have an intensification in spawning in September; in October, a drop in the mean GSI values occurred, and the highest frequencies of post-ovulatory follicles (POFs) were observed. We observed a positive correlation between the POF and the levels of 17α-hydroxyproges- terone. Metynnis maculatus has the potential to be used as a source of pituitary tissue for the preparation of crude extracts for hormonal induction; the theoretical period for use is from September to December, but specific studies to determine the feasibility of this approach must be conducted. Neste estudo, avaliamos a dinâmica da maturação ovariana a desova durante o ciclo reprodutivo de Metynnis maculatus. Fêmeas adultas (n = 36) foram coletadas bimestralmente entre abril de 2010 e março de 2011. -

(Characiformes: Serrasalmidae: Myloplus) from the Brazilian Amazon
Neotropical Ichthyology Original article https://doi.org/10.1590/1982-0224-20190112 urn:lsid:zoobank.org:pub:D73103DD-29FA-4B78-89AE-91FA718A1001 Integrative taxonomy reveals a new species of pacu (Characiformes: Serrasalmidae: Myloplus) from the Brazilian Amazon Rafaela Priscila Ota1, Valéria Nogueira Machado2, Correspondence: Marcelo C. Andrade3, Rupert A. Collins4, Izeni Pires Farias2 Rafaela Priscila Ota 2 [email protected] and Tomas Hrbek Pacus of the genus Myloplus represent a formidable taxonomic challenge, and particularly so for the case of M. asterias and M. rubripinnis, two widespread and common species that harbor considerable morphological diversity. Here we apply DNA barcoding and multiple species discovery methods to find candidate species in this complex group. We report on one well-supported lineage that is also morphologically and ecologically distinct. This lineage represents a new species that can be distinguished from congeners by the presence of dark chromatophores on lateral-line scales, which gives the appearance of a black lateral line. It can be further diagnosed by having 25–29 branched dorsal-fin rays (vs. 18–24), 89–114 perforated scales from the supracleithrum to the end of hypural plate (vs. 56–89), and 98–120 total lateral line scales (vs. 59–97). The new species is widely distributed in the Amazon basin, but seems to have a preference for black- and clearwater habitats. This ecological preference and black lateral line color pattern bears a striking similarity to the recently described silver dollar Submitted September 24, 2019 Metynnis melanogrammus. Accepted February 13, 2020 by George Mattox Keywords: COI gene, Cryptic species, Myloplus asterias, Myloplus rubripinnis, Published April 20, 2020 Neotropical. -

Happy New Year 2015
QUATICAQU AT H E O N - L I N E J O U R N A L O F T H E B R O O K L Y N A Q U A R I U M S O C I E T Y VOL. 28 JANUARY ~ FEBRUARY 2015 N o. 3 Metynnis argenteus Silver Dollar HA PPY NEW YEAR 1 104 Y EARS OF E DUCATING A QUARISTS AQUATICA VOL. 28 JANUARY - FEBRUARY 2015 NO. 3 C ONTENT S PAGE 2 THE AQUATICA STAFF. PAGE 23 NOTABLE NATIVES. All about some of the beautiful North PAGE 3 CALENDAR OF EVENTS. American aquarium fish, seldom seen BAS Events for the years 2015 - 2016 and almost never available commercially. ANTHONY P. KROEGER, BAS PAGE 4 MOLLIES LOVE CRACKERS! Collecting wild Sailfin Mollies in Florida. PAGE 25 SPECIES PROFILE. ANTHONY P. KROEGER, BAS Etheostoma caeruieum , Rainbow Darter. JOHN TODARO, BAS PAGE 6 SPECIES PROFILE. The Sailfin PAGE 26 HOBBY HAPPENINGS. Mollie, Poecili latipinna . JOHN TODARO, BAS The further aquatic adventures of Larry Jinks. PAGE 7 TERRORS OF THE LARRY JINKS, BAS, RAS, NJAS PLANTED AQUARIUM. Keeping Silver dollar fish; you must keep in PAGE 28 CATFISH CONNECTIONS. Sy introduces us to Australia’s yellow mind they’re in the same family as the tandanus. Piranha and are voracious plant eaters. fin JOHN TODARO, BAS SY ANGELICUS, BAS PAGE 10 SPECIES PROFILE. The Silver Dollar, PAGE 29 BLUE VELVET SHRIMP. Another article Metynnis ar genteus . on keeping freshwater shrimp, with information on JOHN TODARO, BAS keeping them healthy. BRAD KEMP, BAS, THE SHRIMP FARM.COM PAGE 11 SAND LOACHES - THEY BREED BY THEMSELVES . -

A Rapid Biological Assessment of the Upper Palumeu River Watershed (Grensgebergte and Kasikasima) of Southeastern Suriname
Rapid Assessment Program A Rapid Biological Assessment of the Upper Palumeu River Watershed (Grensgebergte and Kasikasima) of Southeastern Suriname Editors: Leeanne E. Alonso and Trond H. Larsen 67 CONSERVATION INTERNATIONAL - SURINAME CONSERVATION INTERNATIONAL GLOBAL WILDLIFE CONSERVATION ANTON DE KOM UNIVERSITY OF SURINAME THE SURINAME FOREST SERVICE (LBB) NATURE CONSERVATION DIVISION (NB) FOUNDATION FOR FOREST MANAGEMENT AND PRODUCTION CONTROL (SBB) SURINAME CONSERVATION FOUNDATION THE HARBERS FAMILY FOUNDATION Rapid Assessment Program A Rapid Biological Assessment of the Upper Palumeu River Watershed RAP (Grensgebergte and Kasikasima) of Southeastern Suriname Bulletin of Biological Assessment 67 Editors: Leeanne E. Alonso and Trond H. Larsen CONSERVATION INTERNATIONAL - SURINAME CONSERVATION INTERNATIONAL GLOBAL WILDLIFE CONSERVATION ANTON DE KOM UNIVERSITY OF SURINAME THE SURINAME FOREST SERVICE (LBB) NATURE CONSERVATION DIVISION (NB) FOUNDATION FOR FOREST MANAGEMENT AND PRODUCTION CONTROL (SBB) SURINAME CONSERVATION FOUNDATION THE HARBERS FAMILY FOUNDATION The RAP Bulletin of Biological Assessment is published by: Conservation International 2011 Crystal Drive, Suite 500 Arlington, VA USA 22202 Tel : +1 703-341-2400 www.conservation.org Cover photos: The RAP team surveyed the Grensgebergte Mountains and Upper Palumeu Watershed, as well as the Middle Palumeu River and Kasikasima Mountains visible here. Freshwater resources originating here are vital for all of Suriname. (T. Larsen) Glass frogs (Hyalinobatrachium cf. taylori) lay their -

The Role of Piscivores in a Species-Rich Tropical Food
THE ROLE OF PISCIVORES IN A SPECIES-RICH TROPICAL RIVER A Dissertation by CRAIG ANTHONY LAYMAN Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY August 2004 Major Subject: Wildlife and Fisheries Sciences THE ROLE OF PISCIVORES IN A SPECIES-RICH TROPICAL RIVER A Dissertation by CRAIG ANTHONY LAYMAN Submitted to Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Approved as to style and content by: _________________________ _________________________ Kirk O. Winemiller Lee Fitzgerald (Chair of Committee) (Member) _________________________ _________________________ Kevin Heinz Daniel L. Roelke (Member) (Member) _________________________ Robert D. Brown (Head of Department) August 2004 Major Subject: Wildlife and Fisheries Sciences iii ABSTRACT The Role of Piscivores in a Species-Rich Tropical River. (August 2004) Craig Anthony Layman, B.S., University of Virginia; M.S., University of Virginia Chair of Advisory Committee: Dr. Kirk O. Winemiller Much of the world’s species diversity is located in tropical and sub-tropical ecosystems, and a better understanding of the ecology of these systems is necessary to stem biodiversity loss and assess community- and ecosystem-level responses to anthropogenic impacts. In this dissertation, I endeavored to broaden our understanding of complex ecosystems through research conducted on the Cinaruco River, a floodplain river in Venezuela, with specific emphasis on how a human-induced perturbation, commercial netting activity, may affect food web structure and function. I employed two approaches in this work: (1) comparative analyses based on descriptive food web characteristics, and (2) experimental manipulations within important food web modules. -

Advances in Fish Biology Symposium,” We Are Including 48 Oral and Poster Papers on a Diverse Range of Species, Covering a Number of Topics
Advances in Fish Biology SYMPOSIUM PROCEEDINGS Adalberto Val Don MacKinlay International Congress on the Biology of Fish Tropical Hotel Resort, Manaus Brazil, August 1-5, 2004 Copyright © 2004 Physiology Section, American Fisheries Society All rights reserved International Standard Book Number(ISBN) 1-894337-44-1 Notice This publication is made up of a combination of extended abstracts and full papers, submitted by the authors without peer review. The formatting has been edited but the content is the responsibility of the authors. The papers in this volume should not be cited as primary literature. The Physiology Section of the American Fisheries Society offers this compilation of papers in the interests of information exchange only, and makes no claim as to the validity of the conclusions or recommendations presented in the papers. For copies of these Symposium Proceedings, or the other 20 Proceedings in the Congress series, contact: Don MacKinlay, SEP DFO, 401 Burrard St Vancouver BC V6C 3S4 Canada Phone: 604-666-3520 Fax 604-666-0417 E-mail: [email protected] Website: www.fishbiologycongress.org ii PREFACE Fish are so important in our lives that they have been used in thousands of different laboratories worldwide to understand and protect our environment; to understand and ascertain the foundation of vertebrate evolution; to understand and recount the history of vertebrate colonization of isolated pristine environments; and to understand the adaptive mechanisms to extreme environmental conditions. More importantly, fish are one of the most important sources of protein for the human kind. Efforts at all levels have been made to increase fish production and, undoubtedly, the biology of fish, especially the biology of unknown species, has much to contribute.