Mitosomes in Entamoeba Histolytica Contain a Sulfate Activation Pathway
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anti-Rab11 Antibody (ARG41900)
Product datasheet [email protected] ARG41900 Package: 100 μg anti-Rab11 antibody Store at: -20°C Summary Product Description Goat Polyclonal antibody recognizes Rab11 Tested Reactivity Hu, Ms, Rat, Dog, Mk Tested Application IHC-Fr, IHC-P, WB Host Goat Clonality Polyclonal Isotype IgG Target Name Rab11 Antigen Species Mouse Immunogen Purified recombinant peptides within aa. 110 to the C-terminus of Mouse Rab11a, Rab11b and Rab11c (Rab25). Conjugation Un-conjugated Alternate Names RAB11A: Rab-11; Ras-related protein Rab-11A; YL8 RAB11B: GTP-binding protein YPT3; H-YPT3; Ras-related protein Rab-11B RAB25: RAB11C; CATX-8; Ras-related protein Rab-25 Application Instructions Application table Application Dilution IHC-Fr 1:100 - 1:400 IHC-P 1:100 - 1:400 WB 1:250 - 1:2000 Application Note IHC-P: Antigen Retrieval: Heat mediation was recommended. * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control Hepa cell lysate Calculated Mw 24 kDa Observed Size ~ 26 kDa Properties Form Liquid Purification Affinity purification with immunogen. Buffer PBS, 0.05% Sodium azide and 20% Glycerol. Preservative 0.05% Sodium azide www.arigobio.com 1/3 Stabilizer 20% Glycerol Concentration 3 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. -

Four-Dimensional Live Imaging of Apical Biosynthetic Trafficking Reveals a Post-Golgi Sorting Role of Apical Endosomal Intermediates
Four-dimensional live imaging of apical biosynthetic trafficking reveals a post-Golgi sorting role of apical endosomal intermediates Roland Thuenauera,b,1,2, Ya-Chu Hsua, Jose Maria Carvajal-Gonzaleza,3, Sylvie Debordea,4, Jen-Zen Chuanga, Winfried Römerc,d, Alois Sonnleitnerb, Enrique Rodriguez-Boulana,5, and Ching-Hwa Sunga,5 aMargaret M. Dyson Vision Research Institute, Weill Medical College of Cornell University, New York, NY 10065; bCenter for Advanced Bioanalysis Linz, 4020 Linz, Austria; and cInstitute of Biology II, and dBIOSS Centre for Biological Signalling Studies, Albert-Ludwigs-University Freiburg, 79104 Freiburg, Germany Edited by Keith E. Mostov, University of California School of Medicine, San Francisco, CA, and accepted by the Editorial Board January 17, 2014 (received for review March 11, 2013) Emerging data suggest that in polarized epithelial cells newly is an important regulator of biological processes that require synthesized apical and basolateral plasma membrane proteins apical trafficking, e.g., lumen formation during epithelial tubu- traffic through different endosomal compartments en route to the logenesis (11), apical secretion of discoidal/fusiform vesicles in respective cell surface. However, direct evidence for trans-endo- bladder umbrella cells (12), and apical microvillus morphogenesis somal pathways of plasma membrane proteins is still missing and and rhodopsin localization in fly photoreceptors (13). However, the mechanisms involved are poorly understood. Here, we imaged despite the physiological importance of trans-endosomal traf- the entire biosynthetic route of rhodopsin-GFP, an apical marker in ficking, the underlying mechanisms remain largely unclear. epithelial cells, synchronized through recombinant conditional ag- Previous studies on trans-endosomal trafficking in polarized gregation domains, in live Madin-Darby canine kidney cells using epithelial cells have relied on pulse chase/cell fractionation pro- spinning disk confocal microscopy. -
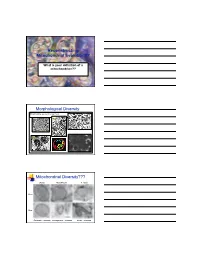
Reconstructing Mitochondrial Evolution?? Morphological Diversity Mitochondrial Diversity???
Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial Diversity??? Chicken Neocallimastix T. foetus 100 nm 50 nm Entamoeba - mitosome microsporidian - mitosome Giardia - mitosome Reconstructing Evolution Mitochondrial evolution well established endosymbiotic theory α-proteobacterium - Rickettsia prowazekii Hydrogenosomal evolution No DNA NOW 2 examples Nyctotherus and Blastocystis (MLO) Several proteins similar to mitochondria Mitosome evolution No DNA Few proteins identified similar to mitochondria Origins via Endosymbiosis Current dogma - mitochondria and related Aerobic α-proteobacterium prokaryote gave rise to present day mitochondria. organelles arose just once in evolution Are hydrogenosomes and mitosomes of anaerobic protists derived from the same proto-mitochondrion? Evidence for: accumulating evidence for several proteins that are currently found in mitochondria - Proteins of Fe-S cluster formation. Scenario A Common ancestor facultative Scenario B organism? degenerate mitochondrion invoke lateral gene transfer from anaerobic prokaryotes Hypotheses for Mito Acquisition Mitochondrion-related Organelles 2 way to interpret this summary - can you think of both? Groups with mitochondrial homologues Hjort et al Phil. Trans. R. Soc. B 2010 365, 713-727 Origin of Hydrogenosomes (Mitosomes?) 1. a)Conversion of Mitochondria HYD b)Common ancestor with mitochondria ? HYD 2. Independent Origin α-proteobacterium ? - anaerobic bacterium HYD Organelles - origins and biogenesis Approaches: (1) Conduct phylogenetic analyses of similar proteins Hsp70 Fd Hsp60 Isc subunits (2) Examine protein targeting to the organelle matrix protein targeting membrane protein targeting (3) Characterize membrane/translocation components These components could have evolved as the endosymbiont was converted to organelle. Reveals evolutionary history. -

A Novel ADP/ATP Transporter in the Mitosome of the Microaerophilic Human Parasite Entamoeba Histolytica
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology, Vol. 15, 737–742, April 26, 2005, ©2005 Elsevier Ltd All rights reserved. DOI 10.1016/j.cub.2005.02.068 A Novel ADP/ATP Transporter in the Mitosome of the Microaerophilic Human Parasite Entamoeba histolytica Ka Wai Chan,1,5 Dirk-Jan Slotboom,1,5 Sian Cox,3 ses confirm that the Entamoeba ADP/ATP carrier is T. Martin Embley,2,* Olivier Fabre,1 distinct from archetypal mitochondrial ADP/ATP carri- Mark van der Giezen,3,6 Marilyn Harding,1 ers, an observation that is supported by its different David S. Horner,4 Edmund R.S. Kunji,1,* substrate and inhibitor specificity. Because many Gloria León-Avila,3,7 and Jorge Tovar3 functions of yeast and human mitochondria rely on 1Dunn Human Nutrition Unit solutes transported by specialized members of this Medical Research Council family, the Entamoeba mitosome must contain only Hills Road a small subset of these processes requiring adenine Cambridge CB2 2XY nucleotide exchange. United Kingdom 2 School of Biology The Devonshire Building Results and Discussion University of Newcastle upon Tyne Newcastle NE1 7RU Identification of an MCF Homolog United Kingdom on the Entamoeba histolytica Genome 3 School of Biological Sciences We identified a homolog of the mitochondrial carrier Royal Holloway family (MCF) on the Entamoeba histolytica genome (TIGR) University of London with BLAST searches. The translated protein contains a Egham, Surrey TW20 0EX tripartite structure [8], signature motifs (IPR001993), and United Kingdom other features typical of MCF members (Figure 1A). -
Drosophila and Human Transcriptomic Data Mining Provides Evidence for Therapeutic
Drosophila and human transcriptomic data mining provides evidence for therapeutic mechanism of pentylenetetrazole in Down syndrome Author Abhay Sharma Institute of Genomics and Integrative Biology Council of Scientific and Industrial Research Delhi University Campus, Mall Road Delhi 110007, India Tel: +91-11-27666156, Fax: +91-11-27662407 Email: [email protected] Nature Precedings : hdl:10101/npre.2010.4330.1 Posted 5 Apr 2010 Running head: Pentylenetetrazole mechanism in Down syndrome 1 Abstract Pentylenetetrazole (PTZ) has recently been found to ameliorate cognitive impairment in rodent models of Down syndrome (DS). The mechanism underlying PTZ’s therapeutic effect is however not clear. Microarray profiling has previously reported differential expression of genes in DS. No mammalian transcriptomic data on PTZ treatment however exists. Nevertheless, a Drosophila model inspired by rodent models of PTZ induced kindling plasticity has recently been described. Microarray profiling has shown PTZ’s downregulatory effect on gene expression in fly heads. In a comparative transcriptomics approach, I have analyzed the available microarray data in order to identify potential mechanism of PTZ action in DS. I find that transcriptomic correlates of chronic PTZ in Drosophila and DS counteract each other. A significant enrichment is observed between PTZ downregulated and DS upregulated genes, and a significant depletion between PTZ downregulated and DS dowwnregulated genes. Further, the common genes in PTZ Nature Precedings : hdl:10101/npre.2010.4330.1 Posted 5 Apr 2010 downregulated and DS upregulated sets show enrichment for MAP kinase pathway. My analysis suggests that downregulation of MAP kinase pathway may mediate therapeutic effect of PTZ in DS. Existing evidence implicating MAP kinase pathway in DS supports this observation. -

A Novel Rab11-Rab3a Cascade Required for Lysosome Exocytosis
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.06.434066; this version posted March 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. A novel Rab11-Rab3a cascade required for lysosome exocytosis Cristina Escrevente1,*, Liliana Bento-Lopes1*, José S Ramalho1, Duarte C Barral1,† 1 iNOVA4Health, CDOC, NOVA Medical School, NMS, Universidade NOVA de Lisboa, 1169-056 Lisboa, Portugal. * These authors contributed equally to this work. † Correspondence should be sent to: Duarte C Barral, CEDOC, NOVA Medical School|Faculdade de Ciências Médicas, Universidade NOVA de Lisboa, Campo dos Mártires da Pátria 130, 1169-056, Lisboa, Portugal, Tel: +351 218 803 102, Fax: +351 218 803 006, [email protected]. (ORCID 0000-0001-8867-2407). Abbreviations used in this paper: FIP, Rab11-family of interacting protein; GEF, guanine nucleotide exchange factor; LE, late endosomes; LRO, lysosome-related organelle; NMIIA, non-muscle myosin heavy chain IIA; Slp-4a, synaptotagmin-like protein 4a. 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.06.434066; this version posted March 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Lysosomes are dynamic organelles, capable of undergoing exocytosis. This process is crucial for several cellular functions, namely plasma membrane repair. -

Bioinformatics Analysis of Autophagy-Related Lncrnas in Esophageal Carcinoma
Bioinformatics analysis of autophagy-related lncRNAs in esophageal carcinoma Dan Wu Shanxi Medical University Yi Ding Xinjiang Medical University junbai fan ( [email protected] ) Shanxi Medical University https://orcid.org/0000-0003-3132-1715 Research article Keywords: Autophagy, Bioinformatics analysis, Esophageal carcinoma Posted Date: November 17th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-60567/v2 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at Combinatorial Chemistry & High Throughput Screening on June 24th, 2021. See the published version at https://doi.org/10.2174/1386207324666210624143452. Page 1/19 Abstract Background: Esophageal carcinoma (ESCA) is a malignant tumor with high invasiveness and mortality. Autophagy has multiple roles in the development of cancer; however, there are limited data on autophagy genes associated with long non-coding RNAs (lncRNAs) in ESCA. The purpose of this study was to screen potential diagnostic and prognostic molecules, and to identify gene co-expression networks associated with autophagy in ESCA. Methods: We downloaded transcriptome expression proles from The Cancer Genome Atlas and autophagy-related gene data from the Human Autophagy Database, and analyzed the co-expression of mRNAs and lncRNAs. In addition, the diagnostic and prognostic value of autophagy-related lncRNAs was analyzed by multivariate Cox regression. Furthermore, Kyoto Encyclopedia of Genes and Genomes analysis was carried out for high-risk patients, and enriched pathways were analyzed by gene set enrichment analysis. Results: The results showed that genes of high-risk patients were enriched in protein export and spliceosome. -

Protein Targeting in Parasites with Cryptic Mitochondria
International Journal for Parasitology 37 (2007) 265–272 www.elsevier.com/locate/ijpara Invited review Protein targeting in parasites with cryptic mitochondria Lena Burri, Patrick J. Keeling * Canadian Institute for Advanced Research, Department of Botany, University of British Columbia, 3529-6270 University Boulevard, Vancouver, BC, Canada V6T 1Z4 Received 4 October 2006; received in revised form 5 December 2006; accepted 11 December 2006 Abstract Many highly specialised parasites have adapted to their environments by simplifying different aspects of their morphology or bio- chemistry. One interesting case is the mitochondrion, which has been subject to strong reductive evolution in parallel in several different parasitic groups. In extreme cases, mitochondria have degenerated so much in physical size and functional complexity that they were not immediately recognised as mitochondria, and are now referred to as ‘cryptic’. Cryptic mitochondrion-derived organelles can be classified as either hydrogenosomes or mitosomes. In nearly all cases they lack a genome and all organellar proteins are nucleus-encoded and expressed in the cytosol. The same is true for the majority of proteins in canonical mitochondria, where the proteins are directed to the organelle by specific targeting sequences (transit peptides) that are recognised by translocases in the mitochondrial membrane. In this review, we compare targeting sequences of different parasitic systems with highly reduced mitochondria and give an overview of how the import machinery has been modified -

Why Chloroplasts and Mitochondria Retain Their Own Genomes and Genetic Systems
PAPER Why chloroplasts and mitochondria retain their own COLLOQUIUM genomes and genetic systems: Colocation for redox regulation of gene expression John F. Allen1 Research Department of Genetics, Evolution and Environment, University College London, London WC1E 6BT, United Kingdom Edited by Patrick J. Keeling, University of British Columbia, Vancouver, BC, Canada, and accepted by the Editorial Board April 26, 2015 (received for review January 1, 2015) Chloroplasts and mitochondria are subcellular bioenergetic organ- control. Fig. 2B illustrates the two possible pathways of synthesis elles with their own genomes and genetic systems. DNA replica- of each of the three token proteins, A, B, and C. Synthesis may tion and transmission to daughter organelles produces cytoplasmic begin with transcription of genes in the endosymbiont or of gene inheritance of characters associated with primary events in photo- copies acquired by the host. CoRR proposes that gene location synthesis and respiration. The prokaryotic ancestors of chloroplasts by itself has no structural or functional consequence for the and mitochondria were endosymbionts whose genes became mature form of any protein whereas natural selection never- copied to the genomes of their cellular hosts. These copies gave theless operates to determine which of the two copies is retained. Selection favors continuity of redox regulation of gene A, and rise to nuclear chromosomal genes that encode cytosolic proteins ’ and precursor proteins that are synthesized in the cytosol for import this regulation is sufficient to render the host s unregulated copy into the organelle into which the endosymbiont evolved. What redundant. In contrast, there is a selective advantage to location of genes B and C in the genome of the host (5), and thus it is the accounts for the retention of genes for the complete synthesis endosymbiont copies of B and C that become redundant and are within chloroplasts and mitochondria of a tiny minority of their lost. -

Supplementary Table 1: Significantly
Supplementary Table 1: Significantly upregulated genes in the “malignant” cluster of PNETs (p<0.05) Fold change Gene (WDEC/WDET) FEV protein 11.61 adenylate cyclase 2 (brain) 4.47 nuclear receptor subfamily 4, group A, member 2 4.45 growth arrest and DNA-damage-inducible, beta 3.28 plexin B1 3.10 neuronal PAS domain protein 2 2.92 caldesmon 1 2.81 drebrin 1 2.43 hypothetical protein BC013764 2.41 TGFB-induced factor (TALE family homeobox) 2.26 Homo sapiens cDNA FLJ12843 fis, clone NT2RP2003293, mRNA sequence 2.17 nuclear prelamin A recognition factor 2.15 chromobox homolog 4 (Pc class homolog, Drosophila) 2.14 RuvB (E coli homolog)-like 1 [Homo sapiens], mRNA sequence 2.03 latent transforming growth factor beta binding protein 4 2.02 putative glialblastoma cell differentiation-related 1.92 hypothetical protein FLJ10298 1.92 protein tyrosine phosphatase type IVA, member 1 1.90 ADP-ribosylation factor-like 4 1.88 NIMA (never in mitosis gene a)-related kinase 7 1.87 RAB11B, member RAS oncogene family 1.87 myelin transcription factor 1 1.86 centaurin, beta 2 1.83 Homo sapiens cDNA FLJ38575 fis, clone HCHON2007046, mRNA sequence 1.83 SH3 domain binding glutamic acid-rich protein like 3 1.81 immunoglobulin heavy constant gamma 3 (G3m marker) 1.77 chromobox homolog 3 (HP1 gamma homolog, Drosophila) 1.74 basic transcription element binding protein 1 1.73 H1 histone family, member X 1.73 protein phosphatase 2A, regulatory subunit B' (PR 53) 1.73 Homo sapiens cDNA FLJ30096 fis, clone BNGH41000045, mRNA sequence 1.73 KIAA0397 gene product 1.71 OK/SW-CL.4 [Homo sapiens], mRNA sequence 1.69 high-mobility group 20B 1.69 phosphoserine phosphatase 1.68 Homo sapiens mRNA; cDNA DKFZp434B142 (from clone DKFZp434B142), mRNA sequence 1.67 nuclear mitotic apparatus protein 1 1.67 hypothetical protein 628 1.66 PRKC, apoptosis, WT1, regulator 1.64 E3 ubiquitin ligase SMURF1 1.64 chromodomain protein, Y chromosome-like 1.63 hypothetical protein FLJ21839 1.62 hypothetical protein MGC2550 1.62 Suppl. -

ARL13B, PDE6D, and CEP164 Form a Functional Network for INPP5E Ciliary Targeting
ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting Melissa C. Humberta,b, Katie Weihbrechta,b, Charles C. Searbyb,c, Yalan Lid, Robert M. Poped, Val C. Sheffieldb,c, and Seongjin Seoa,1 aDepartment of Ophthalmology and Visual Sciences, bDepartment of Pediatrics, cHoward Hughes Medical Institute, and dProteomics Facility, University of Iowa, Iowa City, IA 52242 Edited by Kathryn V. Anderson, Sloan-Kettering Institute, New York, NY, and approved October 19, 2012 (received for review June 28, 2012) Mutations affecting ciliary components cause a series of related polydactyly, skeletal defects, cleft palate, and cerebral develop- genetic disorders in humans, including nephronophthisis (NPHP), mental defects (11). Inactivation of Inpp5e in adult mice results in Joubert syndrome (JBTS), Meckel-Gruber syndrome (MKS), and Bar- obesity and photoreceptor degeneration. Interestingly, many pro- det-Biedl syndrome (BBS), which are collectively termed “ciliopa- teins that localize to cilia, including INPP5E, RPGR, PDE6 α and thies.” Recent protein–protein interaction studies combined with β subunits, GRK1 (Rhodopsin kinase), and GNGT1 (Transducin γ genetic analyses revealed that ciliopathy-related proteins form sev- chain), are prenylated (either farnesylated or geranylgeranylated), eral functional networks/modules that build and maintain the pri- and mutations in these genes or genes involved in their prenylation mary cilium. However, the precise function of many ciliopathy- (e.g., AIPL1 and RCE1) lead to photoreceptor -

(Niacinamide) to Control Skin Aging and Pigmentation
antioxidants Review Mechanistic Basis and Clinical Evidence for the Applications of Nicotinamide (Niacinamide) to Control Skin Aging and Pigmentation Yong Chool Boo Department of Molecular Medicine, School of Medicine, BK21 Plus KNU Biomedical Convergence Program, Cell and Matrix Research Institute, Kyungpook National University, Daegu 41944, Korea; [email protected]; Tel.: +82-53-420-4946 Abstract: Vitamin B3 (nicotinic acid, niacin) deficiency causes the systemic disease pellagra, which leads to dermatitis, diarrhea, dementia, and possibly death depending on its severity and duration. Vitamin B3 is used in the synthesis of the NAD+ family of coenzymes, contributing to cellular energy metabolism and defense systems. Although nicotinamide (niacinamide) is primarily used as a nutritional supplement for vitamin B3, its pharmaceutical and cosmeceutical uses have been extensively explored. In this review, we discuss the biological activities and cosmeceutical properties of nicotinamide in consideration of its metabolic pathways. Supplementation of nicotinamide restores cellular NAD+ pool and mitochondrial energetics, attenuates oxidative stress and inflammatory response, enhances extracellular matrix and skin barrier, and inhibits the pigmentation process in the skin. Topical treatment of nicotinamide, alone or in combination with other active ingredients, reduces the progression of skin aging and hyperpigmentation in clinical trials. Topically applied nicotinamide Citation: Boo, Y.C. Mechanistic Basis and Clinical Evidence for the is well tolerated by the skin. Currently, there is no convincing evidence that nicotinamide has Applications of Nicotinamide specific molecular targets for controlling skin aging and pigmentation. This substance is presumed (Niacinamide) to Control Skin Aging to contribute to maintaining skin homeostasis by regulating the redox status of cells along with and Pigmentation.