Mitochondrion-Related Organelles in Eukaryotic Protists
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The First Cells Were Most Likely Very Simple Prokaryotic Forms. Ra- Spirochetes
T HE O RIGIN OF E UKARYOTIC C ELLS The first cells were most likely very simple prokaryotic forms. Ra- spirochetes. Ingestion of prokaryotes that resembled present-day diometric dating indicates that the earth is 4 to 5 billion years old cyanobacteria could have led to the endosymbiotic development of and that prokaryotes may have arisen more than 3.5 billion years chloroplasts in plants. ago. Eukaryotes are thought to have first appeared about 1.5 billion Another hypothesis for the evolution of eukaryotic cells proposes years ago. that the prokaryotic cell membrane invaginated (folded inward) to en- The eukaryotic cell might have evolved when a large anaerobic close copies of its genetic material (figure 1b). This invagination re- (living without oxygen) amoeboid prokaryote ingested small aerobic (liv- sulted in the formation of several double-membrane-bound entities ing with oxygen) bacteria and stabilized them instead of digesting them. (organelles) in a single cell. These entities could then have evolved This idea is known as the endosymbiont hypothesis (figure 1a) and into the eukaryotic mitochondrion, nucleus, and chloroplasts. was first proposed by Lynn Margulis, a biologist at Boston Univer- Although the exact mechanism for the evolution of the eu- sity. (Symbiosis is an intimate association between two organisms karyotic cell will never be known with certainty, the emergence of of different species.) According to this hypothesis, the aerobic bac- the eukaryotic cell led to a dramatic increase in the complexity and teria developed into mitochondria, which are the sites of aerobic diversity of life-forms on the earth. At first, these newly formed eu- respiration and most energy conversion in eukaryotic cells. -

Bioenergetics and Metabolism Mitochondria Chloroplasts
Bioenergetics and metabolism Mitochondria Chloroplasts Peroxisomes B. Balen Chemiosmosis common pathway of mitochondria, chloroplasts and prokaryotes to harness energy for biological purposes → chemiosmotic coupling – ATP synthesis (chemi) + membrane transport (osmosis) Prokaryotes – plasma membrane → ATP production Eukaryotes – plasma membrane → transport processes – membranes of cell compartments – energy-converting organelles → production of ATP • Mitochondria – fungi, animals, plants • Plastids (chloroplasts) – plants The essential requirements for chemiosmosis source of high-energy e- membrane with embedded proton pump and ATP synthase energy from sunlight or the pump harnesses the energy of e- transfer to pump H+→ oxidation of foodstuffs is proton gradient across the membrane used to create H+ gradient + across a membrane H gradient serves as an energy store that can be used to drive ATP synthesis Figures 14-1; 14-2 Molecular Biology of the Cell (© Garland Science 2008) Electron transport processes (A) mitochondrion converts energy from chemical fuels (B) chloroplast converts energy from sunlight → electron-motive force generated by the 2 photosystems enables the chloroplast to drive electron transfer from H2O to carbohydrate → chloroplast electron transfer is opposite of electron transfer in a mitochondrion Figure 14-3 Molecular Biology of the Cell (© Garland Science 2008) Carbohydrate molecules and O2 are products of the chloroplast and inputs for the mitochondrion Figure 2-41; 2-76 Molecular Biology of the Cell (© Garland -
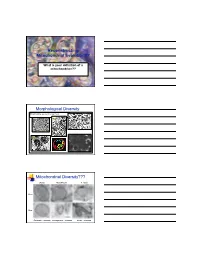
Reconstructing Mitochondrial Evolution?? Morphological Diversity Mitochondrial Diversity???
Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial Diversity??? Chicken Neocallimastix T. foetus 100 nm 50 nm Entamoeba - mitosome microsporidian - mitosome Giardia - mitosome Reconstructing Evolution Mitochondrial evolution well established endosymbiotic theory α-proteobacterium - Rickettsia prowazekii Hydrogenosomal evolution No DNA NOW 2 examples Nyctotherus and Blastocystis (MLO) Several proteins similar to mitochondria Mitosome evolution No DNA Few proteins identified similar to mitochondria Origins via Endosymbiosis Current dogma - mitochondria and related Aerobic α-proteobacterium prokaryote gave rise to present day mitochondria. organelles arose just once in evolution Are hydrogenosomes and mitosomes of anaerobic protists derived from the same proto-mitochondrion? Evidence for: accumulating evidence for several proteins that are currently found in mitochondria - Proteins of Fe-S cluster formation. Scenario A Common ancestor facultative Scenario B organism? degenerate mitochondrion invoke lateral gene transfer from anaerobic prokaryotes Hypotheses for Mito Acquisition Mitochondrion-related Organelles 2 way to interpret this summary - can you think of both? Groups with mitochondrial homologues Hjort et al Phil. Trans. R. Soc. B 2010 365, 713-727 Origin of Hydrogenosomes (Mitosomes?) 1. a)Conversion of Mitochondria HYD b)Common ancestor with mitochondria ? HYD 2. Independent Origin α-proteobacterium ? - anaerobic bacterium HYD Organelles - origins and biogenesis Approaches: (1) Conduct phylogenetic analyses of similar proteins Hsp70 Fd Hsp60 Isc subunits (2) Examine protein targeting to the organelle matrix protein targeting membrane protein targeting (3) Characterize membrane/translocation components These components could have evolved as the endosymbiont was converted to organelle. Reveals evolutionary history. -

Origin and Evolution of Plastids and Mitochondria : the Phylogenetic Diversity of Algae
Cah. Biol. Mar. (2001) 42 : 11-24 Origin and evolution of plastids and mitochondria : the phylogenetic diversity of algae Catherine BOYEN*, Marie-Pierre OUDOT and Susan LOISEAUX-DE GOER UMR 1931 CNRS-Goëmar, Station Biologique CNRS-INSU-Université Paris 6, Place Georges-Teissier, BP 74, F29682 Roscoff Cedex, France. *corresponding author Fax: 33 2 98 29 23 24 ; E-mail: [email protected] Abstract: This review presents an account of the current knowledge concerning the endosymbiotic origin of plastids and mitochondria. The importance of algae as providing a large reservoir of diversified evolutionary models is emphasized. Several reviews describing the plastidial and mitochondrial genome organization and gene content have been published recently. Therefore we provide a survey of the different approaches that are used to investigate the evolution of organellar genomes since the endosymbiotic events. The importance of integrating population genetics concepts to understand better the global evolution of the cytoplasmically inherited organelles is especially emphasized. Résumé : Cette revue fait le point des connaissances actuelles concernant l’origine endosymbiotique des plastes et des mito- chondries en insistant plus particulièrement sur les données portant sur les algues. Ces organismes représentent en effet des lignées eucaryotiques indépendantes très diverses, et constituent ainsi un abondant réservoir de modèles évolutifs. L’organisation et le contenu en gènes des génomes plastidiaux et mitochondriaux chez les eucaryotes ont été détaillés exhaustivement dans plusieurs revues récentes. Nous présentons donc une synthèse des différentes approches utilisées pour comprendre l’évolution de ces génomes organitiques depuis l’événement endosymbiotique. En particulier nous soulignons l’importance des concepts de la génétique des populations pour mieux comprendre l’évolution des génomes à transmission cytoplasmique dans la cellule eucaryote. -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

Localization of the Mitochondrial Ftsz Protein in a Dividing Mitochondrion Mitochondria Are Ubiquitous Organelles That Play Crit
C2001 The Japan Mendel Society Cytologia 66: 421-425, 2001 Localization of the Mitochondrial FtsZ Protein in a Dividing Mitochondrion Manabu Takahara*, Haruko Kuroiwa, Shin-ya Miyagishima, Toshiyuki Mori and Tsuneyoshi Kuroiwa Department of Biological Sciences, Graduate School of Science, University of Tokyo, Hongo, Tokyo 113-0033, Japan Accepted November 2, 2001 Summary FtsZ protein is essential for bacterial cell division, and is also involved in plastid and mitochondrial division. However, little is known of the function of FtsZ in the mitochondrial division process. Here, using electron microscopy, we revealed that the mitochondrial FtsZ (CmFtsZ 1) local- izes at the constricted isthmus of a dividing mitochondrion on the inner (matrix-side) surface of the mitochondrion. These results strongly suggest that the mitochondrial FtsZ acts as a ring structure on the inner surface of mitochondria. Key words Cyanidionschyzon merolae, FtsZ, FtsZ ring, mitochondria, mitochondrion-dividing ring (MD ring) . Mitochondria are ubiquitous organelles that play critical roles in respiration and ATP synthesis in almost all eukaryotic cells. Mitochondria are thought to have originated from a-proteobacteria through an endosymbiont event. Like bacteria, they multiply by the binary fission of pre-existing mitochondria. Although the role of mitochondria in respiration and ATP production is well under- stood, little is known about the proliferation of mitochondria in cells. In plastids, which also arose from prokaryotic endosymbionts, the ring structure that appears at the constricted isthmus of a dividing plastid (the plastid-dividing ring or PD ring) has been iden- tified as the plastid division apparatus (Mita et al. 1986). The PD ring is widespread among plants and algae, and consists of an outer (cytosolic) ring and an inner (stromal) ring in most species. -

A True Symbiosis for the Mitochondria Evolution
: O tics pe ge n r A e c n c e e o s i s B Morelli et al, Bioenergetics 2016, 5:2 Bioenergetics: Open Access DOI: 10.4172/2167-7662.1000137 ISSN: 2167-7662 Letter to Editor Open Access A True Symbiosis for the Mitochondria Evolution Alessandro Morelli1* and Camillo Rosano2 1University of Genova, School of Medical and Pharmaceutical Sciences, Department of Pharmacy, Biochemistry Laboratory, Italy 2UO Proteomics, IRCCS AOU San Martino, IST National Institute for Cancer Research, Largo Rosanna Benzi, Genova, Italy *Corresponding author: Alessandro Morelli, University of Genova, School of Medical and Pharmaceutical Sciences, Department of Pharmacy, Biochemistry Laboratory, Italy, Tel: +39 010 3538153; E-mail: [email protected] Rec Date: June 10, 2016; Acc Date: June 22, 2016; Pub Date: June 24, 2016 Copyright: © 2016 Morelli A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Citation: Morelli A, Rosano C (2016) A True Symbiosis for the Mitochondria Evolution. Bioenergetics 5: 228. doi:10.4172/2167-7662.1000137 Introduction assimilated within eukaryotic cells much later than what initially thought [10]. Endosymbiotic theory (or Symbiogenesis) is an evolutionary theory that was initially proposed more than 100 years ago [1] to explain the These revolutionary data together with the hypothesis by our and origins of eukaryotic cells from the prokaryotic ones. This theory others groups about the possible existence of “extra-mitochondrial” postulated that several key organelles of eukaryotes could have been structures in Eukaryotes with OXPHOS and ATP synthesis perfectly originated as a symbiosis between separate organisms. -

The Revised Classification of Eukaryotes
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/231610049 The Revised Classification of Eukaryotes Article in Journal of Eukaryotic Microbiology · September 2012 DOI: 10.1111/j.1550-7408.2012.00644.x · Source: PubMed CITATIONS READS 961 2,825 25 authors, including: Sina M Adl Alastair Simpson University of Saskatchewan Dalhousie University 118 PUBLICATIONS 8,522 CITATIONS 264 PUBLICATIONS 10,739 CITATIONS SEE PROFILE SEE PROFILE Christopher E Lane David Bass University of Rhode Island Natural History Museum, London 82 PUBLICATIONS 6,233 CITATIONS 464 PUBLICATIONS 7,765 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Biodiversity and ecology of soil taste amoeba View project Predator control of diversity View project All content following this page was uploaded by Smirnov Alexey on 25 October 2017. The user has requested enhancement of the downloaded file. The Journal of Published by the International Society of Eukaryotic Microbiology Protistologists J. Eukaryot. Microbiol., 59(5), 2012 pp. 429–493 © 2012 The Author(s) Journal of Eukaryotic Microbiology © 2012 International Society of Protistologists DOI: 10.1111/j.1550-7408.2012.00644.x The Revised Classification of Eukaryotes SINA M. ADL,a,b ALASTAIR G. B. SIMPSON,b CHRISTOPHER E. LANE,c JULIUS LUKESˇ,d DAVID BASS,e SAMUEL S. BOWSER,f MATTHEW W. BROWN,g FABIEN BURKI,h MICAH DUNTHORN,i VLADIMIR HAMPL,j AARON HEISS,b MONA HOPPENRATH,k ENRIQUE LARA,l LINE LE GALL,m DENIS H. LYNN,n,1 HILARY MCMANUS,o EDWARD A. D. -

Trichonympha Cf
MOLECULAR PHYLOGENETICS OF TRICHONYMPHA CF. COLLARIS AND A PUTATIVE PYRSONYMPHID: THE RELEVANCE TO THE ORIGIN OF SEX by JOEL BRYAN DACKS B.Sc. The University of Alberta, 1995 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER'S OF SCIENCE in THE FACULTY OF GRADUATE STUDIES (Department of Zoology) We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA April 1998 © Joel Bryan Dacks, 1998 In presenting this thesis in partial fulfilment of the requirements for an advanced degree at the University of British Columbia, I agree that the Library shall make it freely available for reference and study. I further agree that permission for extensive copying of this thesis for scholarly purposes may be granted by the head of my department or by his or her representatives. It is understood that copying or publication of this thesis for financial gain shall not be allowed without my written permission. Department of ~2—oc)^Oa^ The University of British Columbia Vancouver, Canada Date {X^ZY Z- V. /^P DE-6 (2/88) Abstract Why sex evolved is one of the central questions in evolutionary genetics. To address this question I have undertaken a molecular phylogenetic study of two candidate lineages to determine the first sexual line. In my thesis the hypermastigotes are confirmed as closely related to the trichomonads in the phylum Parabasalia and found to be more deeply divergent than a putative pyrsonymphid. This means that the Parabasalia are the first sexual lineage. From this I go on to infer that the ancestral sexual cycle included facultative sex. -

Author's Manuscript (764.7Kb)
1 BROADLY SAMPLED TREE OF EUKARYOTIC LIFE Broadly Sampled Multigene Analyses Yield a Well-resolved Eukaryotic Tree of Life Laura Wegener Parfrey1†, Jessica Grant2†, Yonas I. Tekle2,6, Erica Lasek-Nesselquist3,4, Hilary G. Morrison3, Mitchell L. Sogin3, David J. Patterson5, Laura A. Katz1,2,* 1Program in Organismic and Evolutionary Biology, University of Massachusetts, 611 North Pleasant Street, Amherst, Massachusetts 01003, USA 2Department of Biological Sciences, Smith College, 44 College Lane, Northampton, Massachusetts 01063, USA 3Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, 7 MBL Street, Woods Hole, Massachusetts 02543, USA 4Department of Ecology and Evolutionary Biology, Brown University, 80 Waterman Street, Providence, Rhode Island 02912, USA 5Biodiversity Informatics Group, Marine Biological Laboratory, 7 MBL Street, Woods Hole, Massachusetts 02543, USA 6Current address: Department of Epidemiology and Public Health, Yale University School of Medicine, New Haven, Connecticut 06520, USA †These authors contributed equally *Corresponding author: L.A.K - [email protected] Phone: 413-585-3825, Fax: 413-585-3786 Keywords: Microbial eukaryotes, supergroups, taxon sampling, Rhizaria, systematic error, Excavata 2 An accurate reconstruction of the eukaryotic tree of life is essential to identify the innovations underlying the diversity of microbial and macroscopic (e.g. plants and animals) eukaryotes. Previous work has divided eukaryotic diversity into a small number of high-level ‘supergroups’, many of which receive strong support in phylogenomic analyses. However, the abundance of data in phylogenomic analyses can lead to highly supported but incorrect relationships due to systematic phylogenetic error. Further, the paucity of major eukaryotic lineages (19 or fewer) included in these genomic studies may exaggerate systematic error and reduces power to evaluate hypotheses. -

Mitosomes in Entamoeba Histolytica Contain a Sulfate Activation Pathway
Mitosomes in Entamoeba histolytica contain a sulfate activation pathway Fumika Mi-ichi, Mohammad Abu Yousuf, Kumiko Nakada-Tsukui, and Tomoyoshi Nozaki1 Department of Parasitology, National Institute of Infectious Diseases, Tokyo 162-8640, Japan Edited by Andrew Roger, Dalhousie University, Halifax, NS, Canada, and accepted by the Editorial Board October 19, 2009 (received for review June 25, 2009) Hydrogenosomes and mitosomes are mitochondrion-related or- and M. balamuthi lack the ISC system, and instead possess the ganelles in anaerobic/microaerophilic eukaryotes with highly re- nitrogen fixation (NIF) system, which is most likely derived from an duced and divergent functions. The full diversity of their content ancestral nitrogen fixing -proteobacterium by lateral gene transfer and function, however, has not been fully determined. To under- (22, 24). Only 5 proteins have been demonstrated in E. histolytica stand the central role of mitosomes in Entamoeba histolytica,a mitosomes: Cpn60 (8–10, 12), Cpn10 (13), mitochondrial Hsp70 parasitic protozoon that causes amoebic dysentery and liver ab- (11, 15), pyridine nucleotide transhydrogenase (PNT) (2, 8), and scesses, we examined the proteomic profile of purified mitosomes. mitochondria carrier family (MCF, ADP/ATP transporter) (14), Using 2 discontinuous Percoll gradient centrifugation and MS and the central role of mitosomes in E. histolytica remains unknown. analysis, we identified 95 putative mitosomal proteins. Immuno- Analysis of the genome of E. histolytica has not revealed any fluorescence assay showed that 3 proteins involved in sulfate additional information regarding the function of mitosomes and activation, ATP sulfurylase, APS kinase, and inorganic pyrophos- thus, a proteomic analysis of mitosomes seems to be the best phatase, as well as sodium/sulfate symporter, involved in sulfate approach to understand its structure and function (1, 2). -

A Novel ADP/ATP Transporter in the Mitosome of the Microaerophilic Human Parasite Entamoeba Histolytica
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology, Vol. 15, 737–742, April 26, 2005, ©2005 Elsevier Ltd All rights reserved. DOI 10.1016/j.cub.2005.02.068 A Novel ADP/ATP Transporter in the Mitosome of the Microaerophilic Human Parasite Entamoeba histolytica Ka Wai Chan,1,5 Dirk-Jan Slotboom,1,5 Sian Cox,3 ses confirm that the Entamoeba ADP/ATP carrier is T. Martin Embley,2,* Olivier Fabre,1 distinct from archetypal mitochondrial ADP/ATP carri- Mark van der Giezen,3,6 Marilyn Harding,1 ers, an observation that is supported by its different David S. Horner,4 Edmund R.S. Kunji,1,* substrate and inhibitor specificity. Because many Gloria León-Avila,3,7 and Jorge Tovar3 functions of yeast and human mitochondria rely on 1Dunn Human Nutrition Unit solutes transported by specialized members of this Medical Research Council family, the Entamoeba mitosome must contain only Hills Road a small subset of these processes requiring adenine Cambridge CB2 2XY nucleotide exchange. United Kingdom 2 School of Biology The Devonshire Building Results and Discussion University of Newcastle upon Tyne Newcastle NE1 7RU Identification of an MCF Homolog United Kingdom on the Entamoeba histolytica Genome 3 School of Biological Sciences We identified a homolog of the mitochondrial carrier Royal Holloway family (MCF) on the Entamoeba histolytica genome (TIGR) University of London with BLAST searches. The translated protein contains a Egham, Surrey TW20 0EX tripartite structure [8], signature motifs (IPR001993), and United Kingdom other features typical of MCF members (Figure 1A).