RPS9 Antibody Cat
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

C8cc08685k1.Pdf
Electronic Supplementary Material (ESI) for ChemComm. This journal is © The Royal Society of Chemistry 2018 Supporting Information Minimalist Linkers Suitable for Irreversible Inhibitors in Simultaneous Proteome Profiling, Live-Cell Imaging and Drug Screening Cuiping Guo,Yu Chang, Xin Wang, Chengqian Zhang, Piliang Hao*, Ke Ding and Zhengqiu Li* School of Pharmacy, Jinan University, Guangzhou, China 510632 *Corresponding author ([email protected]) 1. General Information All chemicals were purchased from commercial vendors and used without further purification, unless indicated otherwise. All reactions requiring anhydrous conditions were carried out under argon or nitrogen atmosphere using oven-dried glassware. AR-grade solvents were used for all reactions. Reaction progress was monitored by TLC on pre-coated silica plates (Merck 60 F254 nm, 0.25 µm) and spots were visualized by UV, iodine or other suitable stains. Flash column chromatography was carried out using silica gel (Qingdao Ocean). All NMR spectra (1H-NMR, 13C-NMR) were recorded on Bruker 300 MHz/400 MHz NMR spectrometers. Chemical shifts were reported in parts per million (ppm) referenced with respect to appropriate internal standards or residual solvent peaks (CDCl3 = 7.26 ppm, DMSO-d6 = 2.50 ppm). The following abbreviations were used in reporting spectra, br s (broad singlet), s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), dd (doublet of doublets). Mass spectra were obtained on Agilent LC-ESI-MS system. All analytical HPLC were carried out on Agilent system. Water with 0.1% TFA and acetonitrile with 0.1% TFA were used as eluents and the flow rate was 0.5 mL/min. -

Reprogramming of Trna Modifications Controls the Oxidative Stress Response by Codon-Biased Translation of Proteins
Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Chan, Clement T.Y. et al. “Reprogramming of tRNA Modifications Controls the Oxidative Stress Response by Codon-biased Translation of Proteins.” Nature Communications 3 (2012): 937. As Published http://dx.doi.org/10.1038/ncomms1938 Publisher Nature Publishing Group Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/76775 Terms of Use Article is made available in accordance with the publisher's policy and may be subject to US copyright law. Please refer to the publisher's site for terms of use. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins Clement T.Y. Chan,1,2 Yan Ling Joy Pang,1 Wenjun Deng,1 I. Ramesh Babu,1 Madhu Dyavaiah,3 Thomas J. Begley3 and Peter C. Dedon1,4* 1Department of Biological Engineering, 2Department of Chemistry and 4Center for Environmental Health Sciences, Massachusetts Institute of Technology, Cambridge, MA 02139; 3College of Nanoscale Science and Engineering, University at Albany, SUNY, Albany, NY 12203 * Corresponding author: PCD, Department of Biological Engineering, NE47-277, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139; tel 617-253-8017; fax 617-324-7554; email [email protected] 2 ABSTRACT Selective translation of survival proteins is an important facet of cellular stress response. We recently demonstrated that this translational control involves a stress-specific reprogramming of modified ribonucleosides in tRNA. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -
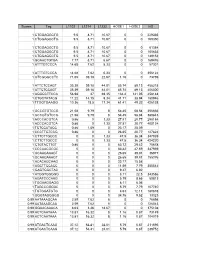
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

Attachment PDF Icon
Spectrum Name of Protein Count of Peptides Ratio (POL2RA/IgG control) POLR2A_228kdBand POLR2A DNA-directed RNA polymerase II subunit RPB1 197 NOT IN CONTROL IP POLR2A_228kdBand POLR2B DNA-directed RNA polymerase II subunit RPB2 146 NOT IN CONTROL IP POLR2A_228kdBand RPAP2 Isoform 1 of RNA polymerase II-associated protein 2 24 NOT IN CONTROL IP POLR2A_228kdBand POLR2G DNA-directed RNA polymerase II subunit RPB7 23 NOT IN CONTROL IP POLR2A_228kdBand POLR2H DNA-directed RNA polymerases I, II, and III subunit RPABC3 19 NOT IN CONTROL IP POLR2A_228kdBand POLR2C DNA-directed RNA polymerase II subunit RPB3 17 NOT IN CONTROL IP POLR2A_228kdBand POLR2J RPB11a protein 7 NOT IN CONTROL IP POLR2A_228kdBand POLR2E DNA-directed RNA polymerases I, II, and III subunit RPABC1 8 NOT IN CONTROL IP POLR2A_228kdBand POLR2I DNA-directed RNA polymerase II subunit RPB9 9 NOT IN CONTROL IP POLR2A_228kdBand ALMS1 ALMS1 3 NOT IN CONTROL IP POLR2A_228kdBand POLR2D DNA-directed RNA polymerase II subunit RPB4 6 NOT IN CONTROL IP POLR2A_228kdBand GRINL1A;Gcom1 Isoform 12 of Protein GRINL1A 6 NOT IN CONTROL IP POLR2A_228kdBand RECQL5 Isoform Beta of ATP-dependent DNA helicase Q5 3 NOT IN CONTROL IP POLR2A_228kdBand POLR2L DNA-directed RNA polymerases I, II, and III subunit RPABC5 5 NOT IN CONTROL IP POLR2A_228kdBand KRT6A Keratin, type II cytoskeletal 6A 3 NOT IN CONTROL IP POLR2A_228kdBand POLR2K DNA-directed RNA polymerases I, II, and III subunit RPABC4 2 NOT IN CONTROL IP POLR2A_228kdBand RFC4 Replication factor C subunit 4 1 NOT IN CONTROL IP POLR2A_228kdBand RFC2 -

The VE-Cadherin/Amotl2 Mechanosensory Pathway Suppresses Aortic In�Ammation and the Formation of Abdominal Aortic Aneurysms
The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inammation and the formation of abdominal aortic aneurysms Yuanyuan Zhang Karolinska Institute Evelyn Hutterer Karolinska Institute Sara Hultin Karolinska Institute Otto Bergman Karolinska Institute Maria Forteza Karolinska Institute Zorana Andonovic Karolinska Institute Daniel Ketelhuth Karolinska University Hospital, Stockholm, Sweden Joy Roy Karolinska Institute Per Eriksson Karolinska Institute Lars Holmgren ( [email protected] ) Karolinska Institute Article Keywords: arterial endothelial cells (ECs), vascular disease, abdominal aortic aneurysms Posted Date: June 15th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-600069/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inflammation and the formation of abdominal aortic aneurysms Yuanyuan Zhang1, Evelyn Hutterer1, Sara Hultin1, Otto Bergman2, Maria J. Forteza2, Zorana Andonovic1, Daniel F.J. Ketelhuth2,3, Joy Roy4, Per Eriksson2 and Lars Holmgren1*. 1Department of Oncology-Pathology, BioClinicum, Karolinska Institutet, Stockholm, Sweden. 2Department of Medicine Solna, BioClinicum, Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden. 3Department of Cardiovascular and Renal Research, Institutet of Molecular Medicine, Univ. of Southern Denmark, Odense, Denmark 4Department of Molecular Medicine and Surgery, Karolinska Institutet, Karolinska University Hospital, Stockholm, -

Supplementary Table 1. the List of Proteins with at Least 2 Unique
Supplementary table 1. The list of proteins with at least 2 unique peptides identified in 3D cultured keratinocytes exposed to UVA (30 J/cm2) or UVB irradiation (60 mJ/cm2) and treated with treated with rutin [25 µM] or/and ascorbic acid [100 µM]. Nr Accession Description 1 A0A024QZN4 Vinculin 2 A0A024QZN9 Voltage-dependent anion channel 2 3 A0A024QZV0 HCG1811539 4 A0A024QZX3 Serpin peptidase inhibitor 5 A0A024QZZ7 Histone H2B 6 A0A024R1A3 Ubiquitin-activating enzyme E1 7 A0A024R1K7 Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein 8 A0A024R280 Phosphoserine aminotransferase 1 9 A0A024R2Q4 Ribosomal protein L15 10 A0A024R321 Filamin B 11 A0A024R382 CNDP dipeptidase 2 12 A0A024R3V9 HCG37498 13 A0A024R3X7 Heat shock 10kDa protein 1 (Chaperonin 10) 14 A0A024R408 Actin related protein 2/3 complex, subunit 2, 15 A0A024R4U3 Tubulin tyrosine ligase-like family 16 A0A024R592 Glucosidase 17 A0A024R5Z8 RAB11A, member RAS oncogene family 18 A0A024R652 Methylenetetrahydrofolate dehydrogenase 19 A0A024R6C9 Dihydrolipoamide S-succinyltransferase 20 A0A024R6D4 Enhancer of rudimentary homolog 21 A0A024R7F7 Transportin 2 22 A0A024R7T3 Heterogeneous nuclear ribonucleoprotein F 23 A0A024R814 Ribosomal protein L7 24 A0A024R872 Chromosome 9 open reading frame 88 25 A0A024R895 SET translocation 26 A0A024R8W0 DEAD (Asp-Glu-Ala-Asp) box polypeptide 48 27 A0A024R9E2 Poly(A) binding protein, cytoplasmic 1 28 A0A024RA28 Heterogeneous nuclear ribonucleoprotein A2/B1 29 A0A024RA52 Proteasome subunit alpha 30 A0A024RAE4 Cell division cycle 42 31 -

Inhibition of the MID1 Protein Complex
Matthes et al. Cell Death Discovery (2018) 4:4 DOI 10.1038/s41420-017-0003-8 Cell Death Discovery ARTICLE Open Access Inhibition of the MID1 protein complex: a novel approach targeting APP protein synthesis Frank Matthes1,MoritzM.Hettich1, Judith Schilling1, Diana Flores-Dominguez1, Nelli Blank1, Thomas Wiglenda2, Alexander Buntru2,HannaWolf1, Stephanie Weber1,InaVorberg 1, Alina Dagane2, Gunnar Dittmar2,3,ErichWanker2, Dan Ehninger1 and Sybille Krauss1 Abstract Alzheimer’s disease (AD) is characterized by two neuropathological hallmarks: senile plaques, which are composed of amyloid-β (Aβ) peptides, and neurofibrillary tangles, which are composed of hyperphosphorylated tau protein. Aβ peptides are derived from sequential proteolytic cleavage of the amyloid precursor protein (APP). In this study, we identified a so far unknown mode of regulation of APP protein synthesis involving the MID1 protein complex: MID1 binds to and regulates the translation of APP mRNA. The underlying mode of action of MID1 involves the mTOR pathway. Thus, inhibition of the MID1 complex reduces the APP protein level in cultures of primary neurons. Based on this, we used one compound that we discovered previously to interfere with the MID1 complex, metformin, for in vivo experiments. Indeed, long-term treatment with metformin decreased APP protein expression levels and consequently Aβ in an AD mouse model. Importantly, we have initiated the metformin treatment late in life, at a time-point where mice were in an already progressed state of the disease, and could observe an improved behavioral phenotype. These 1234567890 1234567890 findings together with our previous observation, showing that inhibition of the MID1 complex by metformin also decreases tau phosphorylation, make the MID1 complex a particularly interesting drug target for treating AD. -

Supplementary Information
1 Supplementary Information 2 PARK2 promotes mitochondrial pathway of apoptosis and 3 antimicrotubule drugs chemosensitivity via degradation of 4 phospho-BCL-2 5 Hengxing Chen1 †, Yun Li1 †, Yu Li1, Zhen Chen1, Limin Xie1, Wenjia Li1, Yuanxin Zhu1, Hong 6 Xue2, H. Phillip Koeffler3,4, Wenjing Wu5, Kaishun Hu1 *, Dong Yin1, * 7 8 Table of contents 9 Supplementary Materials and Methods……………………………………………2 10 Supplementary Figure. 1…………………………………………………………….6 11 Supplementary Figure. 2…………………………………………………………….7 12 Supplementary Figure. 3…………………………………………………………….8 13 Supplementary Figure. 4…………………………………………………………….9 14 Supplementary Figure. 5……………………………………………………………10 15 Supplementary Figure. 6……………………………………………………………11 16 Supplementary Figure. 7……………………………………………………………12 17 Supplementary Table 1……………………………………………...………………13 18 Supplementary Table 2……………………………………………………...………22 19 20 21 22 1 23 24 25 Supplementary Materials and Methods 26 Cell lines 27 The human breast cell lines MCF-7, MDA-MB-231, T47D, ZR-75-30 and MDA-MB-134-VI 28 were obtained from ATCC. MDA-MB-231 were maintained in DMEM supplemented with 29 10% (v/v) FBS and 1% (v/v) Penicillin-Streptomycin, at 37°C, 5% CO2 in a humidified 30 atmosphere. MCF-7, T47D and ZR-75-30 grew in RPMI 1640 with 10% fetal bovine serum 31 and 1% Penicillin-Streptomycin. MDA-MB-134-VI were cultured in Leibovitz's L-15 32 Medium with 20% (v/v) FBS and 1% (v/v) Penicillin-Streptomycin. 33 34 Flow cytometric analysis 35 Cells were treated with designated chemotherapy drugs and then collected for Annexin V and 36 JC-1 analyses. For Annexin V analysis, apoptosis in breast cancer cells was detected using the 37 Annexin V-FITC Apoptosis Detection Kit (Invitrogen) according to the manufacturer's 38 protocol. -

(HBV) Infection of the Liver Ahmed Mohamed Abdel-B Diab Purdue University
Purdue University Purdue e-Pubs Open Access Dissertations Theses and Dissertations January 2016 The oler of PLK1 in Hepatitis B Virus (HBV) infection of the liver Ahmed Mohamed Abdel-B Diab Purdue University Follow this and additional works at: https://docs.lib.purdue.edu/open_access_dissertations Recommended Citation Diab, Ahmed Mohamed Abdel-B, "The or le of PLK1 in Hepatitis B Virus (HBV) infection of the liver" (2016). Open Access Dissertations. 1214. https://docs.lib.purdue.edu/open_access_dissertations/1214 This document has been made available through Purdue e-Pubs, a service of the Purdue University Libraries. Please contact [email protected] for additional information. Graduate School Form 30 Updated 12/26/2015 PURDUE UNIVERSITY GRADUATE SCHOOL Thesis/Dissertation Acceptance This is to certify that the thesis/dissertation prepared By Ahmed Mohamed Abdel-B Diab Entitled The role of PLK1 in Hepatitis B Virus (HBV) Infection of the Liver For the degree of Doctor of Philosophy Is approved by the final examining committee: Ourania Andrisani Andy W. Tao Co-chair Fabien Zoulim Co-chair Robert Geahlen Xiaoqi Liu To the best of my knowledge and as understood by the student in the Thesis/Dissertation Agreement, Publication Delay, and Certification Disclaimer (Graduate School Form 32), this thesis/dissertation adheres to the provisions of Purdue University’s “Policy of Integrity in Research” and the use of copyright material. Approved by Major Professor(s): Ourania Andrisani Laurie Jaeger 10/6/2016 Approved by: Head of the Departmental Graduate Program Date i THE ROLE OF PLK1 IN HEPATITIS B VIRUS (HBV) INFECTION OF THE LIVER A Dissertation Submitted to the Faculty of Purdue University by Ahmed M Diab In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy December 2016 Purdue University West Lafayette, Indiana ii ACKNOWLEDGEMENTS I would like to begin by extending my gratitude to my committee members Robert Geahlen, Andy W. -
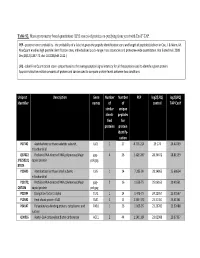
Table S2. Mass Spectrometry Based Quantitation (LFQ Scores) of Proteins Co-Purifying from Yeast with Exoy-TAP
Table S2. Mass spectrometry based quantitation (LFQ scores) of proteins co-purifying from yeast with ExoY-TAP. PEP - posterior error probability - the probability of a false hit given the peptide identification score and length of peptides (defined in Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide quantitation. Nat Biotechnol. 2008 Dec;26(12):1367-72. doi: 10.1038/nbt.1511.) LFQ - Label Free Quantitation score - proportional to the average peptide signal intensity for all the peptides used to identify a given protein. Approximates the relative amounts of protein and can be used to compare protein levels between two conditions. Uniprot Description Gene Number Number PEP log2(LFQ) log2(LFQ) identifier names of of control TAP-ExoY similar unique identi- peptides fied for proteins protein identify- cation P07342 Acetolactate synthase catalytic subunit, ILV2 1 27 8.77E-214 29.174 26.40759 mitochondrial Q87022 Probable RNA-directed RNA polymerase;Major gag- 4 26 1.42E-267 28.34672 28.81559 ;P32503;Q capsid protein pol;gag 87024 P25605 Acetolactate synthase small subunit, ILV6 1 14 7.28E-99 28.14865 25.68654 mitochondrial P23172; Probable RNA-directed RNA polymerase;Major gag- 216 1.63E-75 25.06163 20.40561 Q87026 capsid protein pol;gag P02994 Elongation factor 1-alpha TEF1 1 14 1.47E-75 24.22957 25.30567 P10591 Heat shock protein SSA1 SSA1 2 8 2.54E-173 23.31451 25.41361 P04147 Polyadenylate-binding protein, cytoplasmic and PAB1 126 1.06E-95 23.29782 23.92488 nuclear Q00955 Acetyl-CoA carboxylase;Biotin carboxylase ACC1 2 44 1.54E-164 23.02989 20.97337 P0C2I6;P0 Transposon Ty1-LR3 Gag-Pol polyprotein;Capsid TY1B- 57 3 1.88E-88 22.48395 23.64713 C2I9;P0C2 protein;Ty1 protease;Integrase; Reverse LR3;TY1B- J1;P47098 transcriptase/ribonuclease H (Rev. -

Supplementary Table S5. Protein Expression Changes in the Functionally Annotated Categories Within Cluster from Figure 13A
Supplementary Table S5. Protein expression changes in the functionally annotated categories within cluster from Figure 13A. Human ATPase activity regulation equivalent Accession Zq FDR No. Ac. No. No. Protein 120 1 4 7 120 1 4 7 peptides - I3LJX7 PIG Uncharacterized protein TOR1AIP2 0.01 0.00 0.78 1.03 1 - Q06AA3 PIG Regucalcin 0.03 0.00 0.15 1.00 1 - P09493-6 HUMAN Isoform 6 of Tropomyosin alpha-1 chain 0.13 0.23 0.35 0.97 1 - Q29307 PIG ATPase inhibitor, mitochondrial 0.76 1.01 1.01 0.76 6 - F1S9B6 PIG Uncharacterized protein (Fragment) OXA1L 1.00 0.97 0.74 1.00 1 - P60660 HUMAN Myosin light polypeptide 6 0.07 0.91 1.00 0.64 4 P12829 F1RRT2 PIG Uncharacterized protein (Fragment) MYL4 0.40 1.00 1.01 1.01 3 - A5X497 PIG Cardiac troponin I 1.00 0.97 0.96 1.02 22 P45379 I3LS66 PIG Uncharacterized protein TNNT2 0.94 0.91 1.00 1.02 24 - Q8MKF2 PIG Beta-tropomyosin (Fragment) 1.01 0.88 1.01 1.02 1 - P14632 PIG Lactotransferrin 1.00 0.97 1.00 1.01 2 - P09493-2 HUMAN Isoform 2 of Tropomyosin alpha-1 chain 0.93 0.95 1.00 1.03 1 - Q5JTV8 HUMAN Torsin-1A-interacting protein 1 1.00 0.98 1.01 1.00 1 - P09493-5 HUMAN Isoform 5 of Tropomyosin alpha-1 chain 0.17 0.93 0.95 0.74 1 O95433 F1SE06 PIG Uncharacterized protein LOC100626607 1.00 1.00 1.00 1.02 4 Q01449 F1SSF9 PIG Uncharacterized protein MYL7 0.30 1.00 0.99 0.73 3 - J3KN66 HUMAN Torsin-1A-interacting protein 1 1.00 0.99 0.99 1.02 1 - F1S698 PIG Uncharacterized protein TOR1AIP1 1.00 1.00 0.96 1.00 2 - O95433 HUMAN Activator of 90 kDa heat shock protein ATPase homolog 1 1.00 1.00 1.01 1.00 1 -