Supplementary Table S5. Protein Expression Changes in the Functionally Annotated Categories Within Cluster from Figure 13A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

C8cc08685k1.Pdf
Electronic Supplementary Material (ESI) for ChemComm. This journal is © The Royal Society of Chemistry 2018 Supporting Information Minimalist Linkers Suitable for Irreversible Inhibitors in Simultaneous Proteome Profiling, Live-Cell Imaging and Drug Screening Cuiping Guo,Yu Chang, Xin Wang, Chengqian Zhang, Piliang Hao*, Ke Ding and Zhengqiu Li* School of Pharmacy, Jinan University, Guangzhou, China 510632 *Corresponding author ([email protected]) 1. General Information All chemicals were purchased from commercial vendors and used without further purification, unless indicated otherwise. All reactions requiring anhydrous conditions were carried out under argon or nitrogen atmosphere using oven-dried glassware. AR-grade solvents were used for all reactions. Reaction progress was monitored by TLC on pre-coated silica plates (Merck 60 F254 nm, 0.25 µm) and spots were visualized by UV, iodine or other suitable stains. Flash column chromatography was carried out using silica gel (Qingdao Ocean). All NMR spectra (1H-NMR, 13C-NMR) were recorded on Bruker 300 MHz/400 MHz NMR spectrometers. Chemical shifts were reported in parts per million (ppm) referenced with respect to appropriate internal standards or residual solvent peaks (CDCl3 = 7.26 ppm, DMSO-d6 = 2.50 ppm). The following abbreviations were used in reporting spectra, br s (broad singlet), s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), dd (doublet of doublets). Mass spectra were obtained on Agilent LC-ESI-MS system. All analytical HPLC were carried out on Agilent system. Water with 0.1% TFA and acetonitrile with 0.1% TFA were used as eluents and the flow rate was 0.5 mL/min. -

The Rise and Fall of the Bovine Corpus Luteum
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-6-2017 The Rise and Fall of the Bovine Corpus Luteum Heather Talbott University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Biochemistry Commons, Molecular Biology Commons, and the Obstetrics and Gynecology Commons Recommended Citation Talbott, Heather, "The Rise and Fall of the Bovine Corpus Luteum" (2017). Theses & Dissertations. 207. https://digitalcommons.unmc.edu/etd/207 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. THE RISE AND FALL OF THE BOVINE CORPUS LUTEUM by Heather Talbott A DISSERTATION Presented to the Faculty of the University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biochemistry and Molecular Biology Graduate Program Under the Supervision of Professor John S. Davis University of Nebraska Medical Center Omaha, Nebraska May, 2017 Supervisory Committee: Carol A. Casey, Ph.D. Andrea S. Cupp, Ph.D. Parmender P. Mehta, Ph.D. Justin L. Mott, Ph.D. i ACKNOWLEDGEMENTS This dissertation was supported by the Agriculture and Food Research Initiative from the USDA National Institute of Food and Agriculture (NIFA) Pre-doctoral award; University of Nebraska Medical Center Graduate Student Assistantship; University of Nebraska Medical Center Exceptional Incoming Graduate Student Award; the VA Nebraska-Western Iowa Health Care System Department of Veterans Affairs; and The Olson Center for Women’s Health, Department of Obstetrics and Gynecology, Nebraska Medical Center. -

Reprogramming of Trna Modifications Controls the Oxidative Stress Response by Codon-Biased Translation of Proteins
Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Chan, Clement T.Y. et al. “Reprogramming of tRNA Modifications Controls the Oxidative Stress Response by Codon-biased Translation of Proteins.” Nature Communications 3 (2012): 937. As Published http://dx.doi.org/10.1038/ncomms1938 Publisher Nature Publishing Group Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/76775 Terms of Use Article is made available in accordance with the publisher's policy and may be subject to US copyright law. Please refer to the publisher's site for terms of use. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins Clement T.Y. Chan,1,2 Yan Ling Joy Pang,1 Wenjun Deng,1 I. Ramesh Babu,1 Madhu Dyavaiah,3 Thomas J. Begley3 and Peter C. Dedon1,4* 1Department of Biological Engineering, 2Department of Chemistry and 4Center for Environmental Health Sciences, Massachusetts Institute of Technology, Cambridge, MA 02139; 3College of Nanoscale Science and Engineering, University at Albany, SUNY, Albany, NY 12203 * Corresponding author: PCD, Department of Biological Engineering, NE47-277, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139; tel 617-253-8017; fax 617-324-7554; email [email protected] 2 ABSTRACT Selective translation of survival proteins is an important facet of cellular stress response. We recently demonstrated that this translational control involves a stress-specific reprogramming of modified ribonucleosides in tRNA. -

Protein Name Accession Number Molecular Weight Myovi-GTD
MyoVI-GTD MyoVI-GTD MyoVa-MGT MyoVa-MGT Molecular Spectral Unique Spectral Unique Protein Name Accession Number Weight Counts Peptides Counts Peptides Dync1h1 Cytoplasmic dynein 1 heavy chain 1 IPI00119876 532 kDa 310 121 515 182 Spna2 Spectrin alpha 2 IPI00757353 285 kDa 853 170 597 149 Myo5a 215 kDa protein IPI00875222 215 kDa 162 47 874 109 AU042671 hypothetical protein LOC269700 isoform 1 IPI00762814 453 kDa 2 2 231 104 Spnb2 Isoform 1 of Spectrin beta chain, brain 1 IPI00319830 274 kDa 505 122 347 100 Dmxl2 Isoform 1 of DmX-like protein 2 IPI00853932 338 kDa 63 38 251 100 Cltc Clathrin heavy chain 1 IPI00169916 (+1) 192 kDa 1994 138 565 90 Mtap2 12 days embryo spinal cord cDNA, RIKEN full-length enriched library, clone:C530026F16 product:microtubule-associated protein 2, full insert sequenceIPI00894724 199 kDa 229 82 258 74 Mtap1a Isoform 1 of Microtubule-associated protein 1A IPI00408909 (+1) 300 kDa 310 86 214 74 Itpr1 Isoform 4 of Inositol 1,4,5-trisphosphate receptor type 1 IPI00230019 (+3) 311 kDa 37 18 155 73 Huwe1 HECT, UBA and WWE domain containing 1 IPI00463909 (+1) 483 kDa 5 5 91 69 Fasn Fatty acid synthase IPI00113223 272 kDa 24 17 140 68 Usp9x Ubiquitin carboxyl-terminal hydrolase IPI00798468 291 kDa 68 45 98 65 Lrp1 Prolow-density lipoprotein receptor-related protein 1 precursor IPI00119063 505 kDa 92 53 109 62 Myh10 Myosin-10 IPI00515398 (+1) 229 kDa 65 40 98 59 Mical1 NEDD9-interacting protein with calponin homology and LIM domains IPI00116371 117 kDa 2 2 203 57 Plec1 Isoform PLEC-1I of Plectin-1 IPI00229509 (+10) -

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Immunoprecipitation and Mass Spectrometry Defines an Extensive
BRES : 44759 Model7 pp: À 1221ðcol:fig: : NILÞ brain research ] ( ]]]]) ]]]– ]]] Available online at www.sciencedirect.com 121 122 123 124 125 126 www.elsevier.com/locate/brainres 127 128 129 Review 130 131 fi 132 Immunoprecipitation and mass spectrometry de nes 133 – 134 an extensive RBM45 protein protein interaction 135 Q2 136 network 137 138 a a,b a a c 139 Yang Li , Mahlon Collins , Jiyan An , Rachel Geiser , Tony Tegeler , c c c a,b,n 140 Q1 Kristine Tsantilas , Krystine Garcia , Patrick Pirrotte , Robert Bowser 141 aDivisions of Neurology and Neurobiology, Barrow Neurological Institute, St. Joseph's Hospital and Medical Center, 142 Phoenix, AZ 85013, USA 143 bUniversity of Pittsburgh School of Medicine, Pittsburgh, PA 15261, USA 144 cCenter for Proteomics, TGen (Translational Genomics Research Institute), Phoenix, AZ 85004, USA 145 146 147 article info abstract 148 149 Article history: The pathological accumulation of RNA-binding proteins (RBPs) within inclusion bodies is a 150 Received 30 January 2016 hallmark of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration 151 Received in revised form (FTLD). RBP aggregation results in both toxic gain and loss of normal function. Determining 152 25 February 2016 the protein binding partners and normal functions of disease-associated RBPs is necessary 153 Accepted 28 February 2016 to fully understand molecular mechanisms of RBPs in disease. Herein, we characterized 154 the protein–protein interactions (PPIs) of RBM45, a RBP that localizes to inclusions in ALS/ 155 – fi Keywords: FTLD. Using immunoprecipitation coupled to mass spectrometry (IP MS), we identi ed 132 156 fi RBM45 proteins that speci cally interact with RBM45 within HEK293 cells. -

Profilin-1 Is Required for Survival of Adult Hematopoietic Stem Cells
Extended methods Immunohistochemistry HepG-2, SMMC-7721, and 293T cells were obtained from Cell Resource Center of Shanghai Institute for Biological Science, Chinese Academy Science, Shanghai, China. HUVEC cells were kindly provided by Prof. Ping-Jin Gao at Institute of Health Sciences (Shanghai, China). All these cell lines were cultured in DMEM with 10% FBS. MDA- MB-231 cell line was kindly provided by Prof. Ming-Yao Liu (East China Normal University, Shanghai, China) and was cultured in Leibovitz L-15 medium with 10% FBS. All these cell lines were originally purchased from ATCC. MDA-MB-231, SMMC-7721 or HepG2 cells were grown on coverslips in 24-well plates and fixed in either 4% paraformaldehyde or pre-chilled methanol (-20°C) for 10 min. In some cases, WT or VPS33B-null Lin-Sca-1+c-Kit+Flk2-CD34- LT-HSCs were collected by flow cytometry and fixed for immunofluorescence staining. Cells were then blocked with 3% BSA in PBS for 60 min followed by incubation with primary antibodies overnight. The antibodies used were anti-HA (Sigma), anti-Flag (Sigma), anti-VPS33B (Sigma), anti- VPS16B (Abcam), anti-GDI2 (Proteintech), anti-LAMP1 (Proteintech), anti-FLOT1 (Abways), anti-CD63 (Proteintech), anti-ANGPTL2 (R&D system), anti-ANGPTL3 (R&D system), anti-TPO (Abways), anti-GLUT1 (Proteintech), anti-LDHA (Proteintech), anti-PKM2 (CST), anti-RAB11A (Abways), anti-RAB27A (Abways) and anti-V5 (Biodragon). Fluorescent-conjugated secondary antibodies (Alexa Fluor® 488 or Alexa Fluor® 555) against mouse, rabbit, or goat were obtained from the Thermo Scientific Inc. The details for all the antibodies are listed in Table S3. -

Supplementary Table
Supporting information Additional Supporting Information may be found in the online version of this article: Supplementary Table S1: List of deregulated genes in serum of cancer patients in comparision to serum of healthy individuals (p < 0.05, logFC ≥ 1). ENTREZ Gene ID Symbol Gene Name logFC p-value q-value 8407 TAGLN2 transgelin 2 3,78 3,40E-07 0,0022 7035 TFPI tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) 3,53 4,30E-05 0,022 28996 HIPK2 homeodomain interacting protein kinase 2 3,49 2,50E-06 0,0066 3690 ITGB3 integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) 3,48 0,00053 0,081 7035 TFPI tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) 3,45 1,80E-05 0,014 4900 NRGN neurogranin (protein kinase C substrate, RC3) 3,32 0,00012 0,037 10398 MYL9 myosin, light chain 9, regulatory 3,22 8,20E-06 0,011 3796 KIF2A kinesin heavy chain member 2A 3,14 0,00015 0,04 5476 CTSA cathepsin A 3,08 0,00015 0,04 6648 SOD2 superoxide dismutase 2, mitochondrial 3,07 4,20E-06 0,0077 2982 GUCY1A3 guanylate cyclase 1, soluble, alpha 3 3,07 0,0015 0,13 8459 TPST2 tyrosylprotein sulfotransferase 2 3,05 0,00043 0,074 2983 GUCY1B3 guanylate cyclase 1, soluble, beta 3 3,04 3,70E-05 0,021 145781 GCOM1 GRINL1A complex locus 3,02 0,00027 0,059 10611 PDLIM5 PDZ and LIM domain 5 2,87 1,80E-05 0,014 5567 PRKACB protein kinase, cAMP-dependent, catalytic, beta 2,85 0,0015 0,13 25907 TMEM158 transmembrane protein 158 (gene/pseudogene) 2,84 0,0068 0,27 8848 TSC22D1 TSC22 domain family, member 1 2,83 0,00058 0,084 26 351 APP amyloid beta (A4) precursor protein 2,82 0,00018 0,045 9240 PNMA1 paraneoplastic antigen MA1 2,78 0,00028 0,06 400073 C12orf76 chromosome 12 open reading frame 76 2,78 0,00069 0,091 649260 ILMN_35781 PREDICTED: Homo sapiens similar to LIM and senescent cell antigen-like domains 1 (LOC649260), mRNA. -
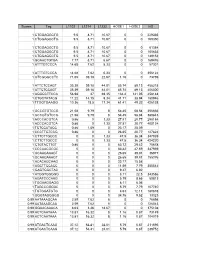
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

Attachment PDF Icon
Spectrum Name of Protein Count of Peptides Ratio (POL2RA/IgG control) POLR2A_228kdBand POLR2A DNA-directed RNA polymerase II subunit RPB1 197 NOT IN CONTROL IP POLR2A_228kdBand POLR2B DNA-directed RNA polymerase II subunit RPB2 146 NOT IN CONTROL IP POLR2A_228kdBand RPAP2 Isoform 1 of RNA polymerase II-associated protein 2 24 NOT IN CONTROL IP POLR2A_228kdBand POLR2G DNA-directed RNA polymerase II subunit RPB7 23 NOT IN CONTROL IP POLR2A_228kdBand POLR2H DNA-directed RNA polymerases I, II, and III subunit RPABC3 19 NOT IN CONTROL IP POLR2A_228kdBand POLR2C DNA-directed RNA polymerase II subunit RPB3 17 NOT IN CONTROL IP POLR2A_228kdBand POLR2J RPB11a protein 7 NOT IN CONTROL IP POLR2A_228kdBand POLR2E DNA-directed RNA polymerases I, II, and III subunit RPABC1 8 NOT IN CONTROL IP POLR2A_228kdBand POLR2I DNA-directed RNA polymerase II subunit RPB9 9 NOT IN CONTROL IP POLR2A_228kdBand ALMS1 ALMS1 3 NOT IN CONTROL IP POLR2A_228kdBand POLR2D DNA-directed RNA polymerase II subunit RPB4 6 NOT IN CONTROL IP POLR2A_228kdBand GRINL1A;Gcom1 Isoform 12 of Protein GRINL1A 6 NOT IN CONTROL IP POLR2A_228kdBand RECQL5 Isoform Beta of ATP-dependent DNA helicase Q5 3 NOT IN CONTROL IP POLR2A_228kdBand POLR2L DNA-directed RNA polymerases I, II, and III subunit RPABC5 5 NOT IN CONTROL IP POLR2A_228kdBand KRT6A Keratin, type II cytoskeletal 6A 3 NOT IN CONTROL IP POLR2A_228kdBand POLR2K DNA-directed RNA polymerases I, II, and III subunit RPABC4 2 NOT IN CONTROL IP POLR2A_228kdBand RFC4 Replication factor C subunit 4 1 NOT IN CONTROL IP POLR2A_228kdBand RFC2 -

The VE-Cadherin/Amotl2 Mechanosensory Pathway Suppresses Aortic In�Ammation and the Formation of Abdominal Aortic Aneurysms
The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inammation and the formation of abdominal aortic aneurysms Yuanyuan Zhang Karolinska Institute Evelyn Hutterer Karolinska Institute Sara Hultin Karolinska Institute Otto Bergman Karolinska Institute Maria Forteza Karolinska Institute Zorana Andonovic Karolinska Institute Daniel Ketelhuth Karolinska University Hospital, Stockholm, Sweden Joy Roy Karolinska Institute Per Eriksson Karolinska Institute Lars Holmgren ( [email protected] ) Karolinska Institute Article Keywords: arterial endothelial cells (ECs), vascular disease, abdominal aortic aneurysms Posted Date: June 15th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-600069/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inflammation and the formation of abdominal aortic aneurysms Yuanyuan Zhang1, Evelyn Hutterer1, Sara Hultin1, Otto Bergman2, Maria J. Forteza2, Zorana Andonovic1, Daniel F.J. Ketelhuth2,3, Joy Roy4, Per Eriksson2 and Lars Holmgren1*. 1Department of Oncology-Pathology, BioClinicum, Karolinska Institutet, Stockholm, Sweden. 2Department of Medicine Solna, BioClinicum, Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden. 3Department of Cardiovascular and Renal Research, Institutet of Molecular Medicine, Univ. of Southern Denmark, Odense, Denmark 4Department of Molecular Medicine and Surgery, Karolinska Institutet, Karolinska University Hospital, Stockholm,