On the Systematics of the Fungus Gnat Subfamily Mycetophilinae (Diptera): a Combined Morphological and Molecular Approach
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -
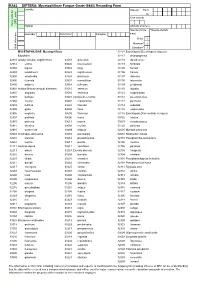
Ra82 Diptera
RA82 DIPTERA: Mycetophilinae Fungus Gnats (6480) Recording Form Locality Date(s) from: to: Vice county GPS users Grey cells for Habitat Altitude (metres) Source (circle *Source details Recorder Determiner Compiler one) Field 1 Museum* 2 Grid reference Literature* 3 MYCETOPHILIDAE: Mycetophilinae 33101 Exechiopsis (Exechiopsis) clypeata Exechiini 33117 dryaspagensis 32801 Allodia (Allodia) anglofennica 33519 griseolum33118 dumitrescae 32912 embla 33526 intermedium33119 fimbriata 32802 lugens 33522 kingi33120 furcata 32803 lundstroemi 33523 nigrofuscum33106 hammi 32804 ornaticollis 33524 proximum33107 indecisa 32806 truncata 33527 rosmellitum33108 intersecta 32805 zaitzevi 33514 ruficorne33109 jenkinsoni 32901 Allodia (Brachycampta) alternans 33515 serenum33110 ligulata 32910 angulata 33516 sericoma33121 magnicauda 32903 barbata 33601 Cordyla brevicornis33112 pseudindecisa 32904 czernyi 33602 crassicornis33113 pulchella 32909 foliifera 33603 fasciata33114 subulata 32905 grata 33604 fissa33115 unguiculata 32906 neglecta 33605 flaviceps33116 Exechiopsis (Xenexechia) crucigera 32907 pistillata 33606 fusca33002 leptura 32915 protenta 33613 insons33003 membranacea 32914 silvatica 33608 murina33122 pollicata 32916 westerholti 33609 nitidula32605 Myrosia maculosa 32602 Allodiopsis domestica 33610 parvipalpis32601 Notolopha cristata 32610 korolevi 33614 pseudomurina32701 Pseudexechia aurivernica 32607 rustica 33611 pusilla32706 monica 32212 Anatella alpina 33612 semiflava32705 parallela 32213 ankeli 33201 Exechia bicincta32703 trisignata 32216 bremia -

R. P. LANE (Department of Entomology), British Museum (Natural History), London SW7 the Diptera of Lundy Have Been Poorly Studied in the Past
Swallow 3 Spotted Flytcatcher 28 *Jackdaw I Pied Flycatcher 5 Blue Tit I Dunnock 2 Wren 2 Meadow Pipit 10 Song Thrush 7 Pied Wagtail 4 Redwing 4 Woodchat Shrike 1 Blackbird 60 Red-backed Shrike 1 Stonechat 2 Starling 15 Redstart 7 Greenfinch 5 Black Redstart I Goldfinch 1 Robin I9 Linnet 8 Grasshopper Warbler 2 Chaffinch 47 Reed Warbler 1 House Sparrow 16 Sedge Warbler 14 *Jackdaw is new to the Lundy ringing list. RECOVERIES OF RINGED BIRDS Guillemot GM I9384 ringed 5.6.67 adult found dead Eastbourne 4.12.76. Guillemot GP 95566 ringed 29.6.73 pullus found dead Woolacombe, Devon 8.6.77 Starling XA 92903 ringed 20.8.76 found dead Werl, West Holtun, West Germany 7.10.77 Willow Warbler 836473 ringed 14.4.77 controlled Portland, Dorset 19.8.77 Linnet KC09559 ringed 20.9.76 controlled St Agnes, Scilly 20.4.77 RINGED STRANGERS ON LUNDY Manx Shearwater F.S 92490 ringed 4.9.74 pullus Skokholm, dead Lundy s. Light 13.5.77 Blackbird 3250.062 ringed 8.9.75 FG Eksel, Belgium, dead Lundy 16.1.77 Willow Warbler 993.086 ringed 19.4.76 adult Calf of Man controlled Lundy 6.4.77 THE DIPTERA (TWO-WINGED FLffiS) OF LUNDY ISLAND R. P. LANE (Department of Entomology), British Museum (Natural History), London SW7 The Diptera of Lundy have been poorly studied in the past. Therefore, it is hoped that the production of an annotated checklist, giving an indication of the habits and general distribution of the species recorded will encourage other entomologists to take an interest in the Diptera of Lundy. -

The Nestling Diet of Svalbard Snow Buntings Identified by DNA Metabarcoding
Faculty of Biosciences, Fisheries and Economics, Department of Arctic and Marine Biology The nestling diet of Svalbard snow buntings identified by DNA metabarcoding — Christian Stolz BIO-3950 Master thesis in Biology, Northern Populations and Ecosystems, May 2019 Faculty of Biosciences, Fisheries and Economics, Department of Arctic and Marine Biology The nestling diet of Svalbard snow buntings identified by DNA metabarcoding Christian Stolz, UiT The Arctic University of Norway, Tromsø, Norway and The University Centre in Svalbard (UNIS), Longyearbyen, Norway BIO-3950 Master Thesis in Biology, Northern Populations and Ecosystems, May 2018 Supervisors: Frode Fossøy, Norwegian Institute for Nature Research (NINA), Trondheim, Norway Øystein Varpe, The University Centre in Svalbard (UNIS), Longyearbyen, Norway Rolf Anker Ims, UiT The Arctic University of Norway, Tromsø, Norway i Abstract Tundra arthropods have considerable ecological importance as a food source for several bird species that are reproducing in the Arctic. The actual arthropod taxa comprising the chick diet are however rarely known, complicating assessments of ecological interactions. In this study, I identified the nestling diet of Svalbard snow bunting (Plectrophenax nivalis) for the first time. Faecal samples of snow bunting chicks were collected in Adventdalen, Svalbard in the breeding season 2018 and analysed via DNA metabarcoding. Simultaneously, the availability of prey arthropods was measured via pitfall trapping. The occurrence of 32 identified prey taxa in the nestling diet changed according to varying abundances and emergence patterns within the tun- dra arthropod community: Snow buntings provisioned their offspring mainly with the most abundant prey items which were in the early season different Chironomidae (Diptera) taxa and Scathophaga furcata (Diptera: Scathophagidae), followed by Spilogona dorsata (Diptera: Mus- cidae). -

Recent Noteworthy Findings of Fungus Gnats from Finland and Northwestern Russia (Diptera: Ditomyiidae, Keroplatidae, Bolitophilidae and Mycetophilidae)
Biodiversity Data Journal 2: e1068 doi: 10.3897/BDJ.2.e1068 Taxonomic paper Recent noteworthy findings of fungus gnats from Finland and northwestern Russia (Diptera: Ditomyiidae, Keroplatidae, Bolitophilidae and Mycetophilidae) Jevgeni Jakovlev†, Jukka Salmela ‡,§, Alexei Polevoi|, Jouni Penttinen ¶, Noora-Annukka Vartija# † Finnish Environment Insitutute, Helsinki, Finland ‡ Metsähallitus (Natural Heritage Services), Rovaniemi, Finland § Zoological Museum, University of Turku, Turku, Finland | Forest Research Institute KarRC RAS, Petrozavodsk, Russia ¶ Metsähallitus (Natural Heritage Services), Jyväskylä, Finland # Toivakka, Myllyntie, Finland Corresponding author: Jukka Salmela ([email protected]) Academic editor: Vladimir Blagoderov Received: 10 Feb 2014 | Accepted: 01 Apr 2014 | Published: 02 Apr 2014 Citation: Jakovlev J, Salmela J, Polevoi A, Penttinen J, Vartija N (2014) Recent noteworthy findings of fungus gnats from Finland and northwestern Russia (Diptera: Ditomyiidae, Keroplatidae, Bolitophilidae and Mycetophilidae). Biodiversity Data Journal 2: e1068. doi: 10.3897/BDJ.2.e1068 Abstract New faunistic data on fungus gnats (Diptera: Sciaroidea excluding Sciaridae) from Finland and NW Russia (Karelia and Murmansk Region) are presented. A total of 64 and 34 species are reported for the first time form Finland and Russian Karelia, respectively. Nine of the species are also new for the European fauna: Mycomya shewelli Väisänen, 1984,M. thula Väisänen, 1984, Acnemia trifida Zaitzev, 1982, Coelosia gracilis Johannsen, 1912, Orfelia krivosheinae Zaitzev, 1994, Mycetophila biformis Maximova, 2002, M. monstera Maximova, 2002, M. uschaica Subbotina & Maximova, 2011 and Trichonta palustris Maximova, 2002. Keywords Sciaroidea, Fennoscandia, faunistics © Jakovlev J et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. -

Mycetophilidae) from Kazakhstan
© Entomologica Fennica. 16 June 2006 Three new species of Docosia Winnertz (Diptera: Mycetophilidae) from Kazakhstan Olavi Kurina Kurina, O. 2006: Three new species of Docosia Winnertz (Diptera: Myceto- philidae) from Kazakhstan. — Entomol. Fennica 17: 110—117. Three new species from Sogety Mountains and Charyn Canyon (Kazakhstan), viz. Docosia selini sp. n., D. agnesiana sp. n. and D. sogetensis sp. n., are de- scribed and detailed illustrations of male terminalia presented. Morphological differences especially in male terminalia are discussed. A key to the Central Asian species ofDocosia is provided. 0. Kurina, Institute ongricultural andEnvironmental Sciences, Estonian Agri— cultural University, Riia st 181, 51014 Tartu, Estonia; E—mail.‘ [email protected] Received 13 April 2005, accepted 22 July 2005 1. Introduction Lastovka. Unfortunately the synopsis compiled by Dr. Lastovka comprising over 100 Holarctic In his monograph, Winnertz (1863) described a species, of which ca. 30 were European, still re- new genus, Docosia, to distinguish two species: mains unpublished (Chandler & Ribeiro 1995). D. sciarina (Meigen, 1830) and D. valida Win- After Dr. Lastovka recently deceased Jan §evcik nertz, 1863. The special review ofthe genus pub- (pers. comm.) will soon finish Lastovka’s revi- lished by Landrock (1916) includes seven spe- sion ofDocosia species of Czech and Slovak Re- cies. Hackman (1988) reported 16 species from publics. the Palaearctic region. Descriptions of ten West- Docosia belongs to the tribe Leiini and, as Palaearctic species have been published subse- suggested by earlier authors, is most closely re- quently by Plassmann (1986, 1996), Chandler lated to Tetragoneura Winnertz, 1846 (e. g. Ed- (1994), Chandler & Ribeiro (1995) and Chandler wards 1925). -

Of Greece, Its Islands
CHANDLERet al.: 255-314 - Studia dipterologica 12 (2005) Heft 2 ISSN 0945-3954 The Fungus Gnats (Diptera: Bolitophilidae, Diadocidiidae, Ditomyiidae , Keroplatidae and Mycetophilidae) of Greece, its islands and Cyprus [Die Pilzmiicken (Diptera: Bolitophilidae, Diadocidiidae, Ditomyiidae, Keroplatidae und Mycetophilidae) Griechenlands und seiner Inseln sowie Zypern4 1 by Peter J. CHANDLER, Dimitar N. BECHEV and Norbert CASPERS Mclksham (UK) Plovdiv (Bulgaria) Bechen (Gernlany) - - -. - ~ Abstract The spccics of fungu\ gnats (Bolitophilidae, Diadoc~dildae,Ditomyiidac. Keroplat~d:~eand Mycetophilidae) o~urringin Greece and Cyprus are reviewed. Altogether 201 species :Ire recorded, 189 for Greece and 69 for Cyprus. Of these 126 specie5 arc newly recorded fol. Greece and 36 arc newly recorded for Cyprus. The following new taxa arc described from Greece: Macrorrhyrtcha ibis spec. nov., M. pelargos spec. nov., M. laconica spec. nov., Macrocera critica spec. nov., Docosia cephaloniae spec. nov., D. enos spec. nov., D. pa- siphae spec. nov., Megophthalmidia illyrica spec. nov.. M. ionica spec. nov., M. pytho spec. nov., Mycomya thrakis spec. nov., Allocolocera scheria spec. nov., Sciophila pandora spec. nov., Ryrnosia labyrinthos spec. nov.; M. ill\,ric,cr is also recorded troln Croc~lia.The follow- ing ncw taa are described from Cyprus: Macrocera cypriaca spec. nov., Megophthalmidia alrzicola spec. nov., M. cedricola spec. nov. The following neu synonymies are propod: M!,c,c~r~iwrenuis I WXLKER,1856) = M. interniissa PL.ASSMA~N,l984 syn. nov., Plrror~rtr~1.illi.s- torri DLIFI>ZICKI,1889 = P rnciscr CASFERS,1991 syn. nov. A key is provided for thc western Palaearctic specie5 of M(ic-i.orrh~~~ic-IrciWI~~ERTZ. -

Zootaxa, the Fungus Gnats (Diptera: Bolitophilidae
Zootaxa 2318: 450–506 (2009) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2009 · Magnolia Press ISSN 1175-5334 (online edition) The fungus gnats (Diptera: Bolitophilidae, Keroplatidae, Mycetophilidae) of Sardinia, with description of six new species* PETER J. CHANDLER 606B Berryfield Lane, Melksham, Wilts SN12 6EL, United Kingdom. E-mail: [email protected] *In: Cerretti, P., Mason, F., Minelli, A., Nardi, G. & Whitmore, D. (Eds), Research on the Terrestrial Arthropods of Sardinia (Italy). Zootaxa, 2318, 1–602. Table of contents Abstract . .450 Introduction . .451 Study area . .452 Material and methods . .452 Abbreviations . .453 Sampling sites . .454 Faunistic list . .456 Bolitophilidae . .456 Keroplatidae, Keroplatinae, Keroplatini . .456 Orfeliini . .457 Macrocerinae . .462 Mycetophilidae, Gnoristinae . .465 Leiinae . .469 Mycetophilinae, Exechiini . .472 Mycetophilini . .480 Mycomyinae . .492 Sciophilinae . .495 Discussion . .500 Acknowledgements . .501 References . .502 Abstract The fungus gnat fauna of Sardinia is reviewed and data presented for all species recorded. Altogether one species of Bolitophilidae, 16 species of Keroplatidae and 105 species of Mycetophilidae are recognised as occurring in Sardinia. As the bolitophilid and two of the mycetophilid species are represented only by females and are not determined to species level, the total confirmed Sardinian list stands at 119 species. Four species of Keroplatidae and 19 species of Mycetophilidae are new to the total Italian fauna, whereas three species of Keroplatidae and 32 species of Mycetophilidae are newly recorded for the island of Sardinia. Six species are described as new to science: two Keroplatidae (Urytalpa juliae sp. nov., Macrocera nuragica sp. nov.) and four Mycetophilidae (Boletina ichnusa sp. nov., Trichonta sandalyon sp. -

Somerset's Ecological Network
Somerset’s Ecological Network Mapping the components of the ecological network in Somerset 2015 Report This report was produced by Michele Bowe, Eleanor Higginson, Jake Chant and Michelle Osbourn of Somerset Wildlife Trust, and Larry Burrows of Somerset County Council, with the support of Dr Kevin Watts of Forest Research. The BEETLE least-cost network model used to produce Somerset’s Ecological Network was developed by Forest Research (Watts et al, 2010). GIS data and mapping was produced with the support of Somerset Environmental Records Centre and First Ecology Somerset Wildlife Trust 34 Wellington Road Taunton TA1 5AW 01823 652 400 Email: [email protected] somersetwildlife.org Front Cover: Broadleaved woodland ecological network in East Mendip Contents 1. Introduction .................................................................................................................... 1 2. Policy and Legislative Background to Ecological Networks ............................................ 3 Introduction ............................................................................................................... 3 Government White Paper on the Natural Environment .............................................. 3 National Planning Policy Framework ......................................................................... 3 The Habitats and Birds Directives ............................................................................. 4 The Conservation of Habitats and Species Regulations 2010 .................................. -

ARTHROPODA Subphylum Hexapoda Protura, Springtails, Diplura, and Insects
NINE Phylum ARTHROPODA SUBPHYLUM HEXAPODA Protura, springtails, Diplura, and insects ROD P. MACFARLANE, PETER A. MADDISON, IAN G. ANDREW, JOCELYN A. BERRY, PETER M. JOHNS, ROBERT J. B. HOARE, MARIE-CLAUDE LARIVIÈRE, PENELOPE GREENSLADE, ROSA C. HENDERSON, COURTenaY N. SMITHERS, RicarDO L. PALMA, JOHN B. WARD, ROBERT L. C. PILGRIM, DaVID R. TOWNS, IAN McLELLAN, DAVID A. J. TEULON, TERRY R. HITCHINGS, VICTOR F. EASTOP, NICHOLAS A. MARTIN, MURRAY J. FLETCHER, MARLON A. W. STUFKENS, PAMELA J. DALE, Daniel BURCKHARDT, THOMAS R. BUCKLEY, STEVEN A. TREWICK defining feature of the Hexapoda, as the name suggests, is six legs. Also, the body comprises a head, thorax, and abdomen. The number A of abdominal segments varies, however; there are only six in the Collembola (springtails), 9–12 in the Protura, and 10 in the Diplura, whereas in all other hexapods there are strictly 11. Insects are now regarded as comprising only those hexapods with 11 abdominal segments. Whereas crustaceans are the dominant group of arthropods in the sea, hexapods prevail on land, in numbers and biomass. Altogether, the Hexapoda constitutes the most diverse group of animals – the estimated number of described species worldwide is just over 900,000, with the beetles (order Coleoptera) comprising more than a third of these. Today, the Hexapoda is considered to contain four classes – the Insecta, and the Protura, Collembola, and Diplura. The latter three classes were formerly allied with the insect orders Archaeognatha (jumping bristletails) and Thysanura (silverfish) as the insect subclass Apterygota (‘wingless’). The Apterygota is now regarded as an artificial assemblage (Bitsch & Bitsch 2000). -

Cambodian Journal of Natural History
Cambodian Journal of Natural History Giant ibis census Patterns of salt lick use Protected area revisions Economic contribution of NTFPs New plants, bees and range extensions June 2016 Vol. 2016 No. 1 Cambodian Journal of Natural History ISSN 2226–969X Editors Email: [email protected] • Dr Neil M. Furey, Chief Editor, Fauna & Flora International, Cambodia. • Dr Jenny C. Daltry, Senior Conservation Biologist, Fauna & Flora International, UK. • Dr Nicholas J. Souter, Mekong Case Study Manager, Conservation International, Cambodia. • Dr Ith Saveng, Project Manager, University Capacity Building Project, Fauna & Flora International, Cambodia. International Editorial Board • Dr Stephen J. Browne, Fauna & Flora International, • Dr Sovanmoly Hul, Muséum National d’Histoire Singapore. Naturelle, Paris, France. • Dr Martin Fisher, Editor of Oryx – The International • Dr Andy L. Maxwell, World Wide Fund for Nature, Journal of Conservation, Cambridge, U.K. Cambodia. • Dr L. Lee Grismer, La Sierra University, California, • Dr Brad Pett itt , Murdoch University, Australia. USA. • Dr Campbell O. Webb, Harvard University Herbaria, • Dr Knud E. Heller, Nykøbing Falster Zoo, Denmark. USA. Other peer reviewers for this volume • Prof. Leonid Averyanov, Komarov Botanical Institute, • Neang Thy, Minstry of Environment, Cambodia. Russia. • Dr Nguyen Quang Truong, Institute of Ecology and • Prof. John Blake, University of Florida, USA. Biological Resources, Vietnam. • Dr Stephan Gale, Kadoorie Farm & Botanic Garden, • Dr Alain Pauly, Royal Belgian Institute of Natural Hong Kong. Sciences, Belgium. • Fredéric Goes, Cambodia Bird News, France. • Dr Colin Pendry, Royal Botanical Garden, Edinburgh, • Dr Hubert Kurzweil, Singapore Botanical Gardens, UK. Singapore. • Dr Stephan Risch, Leverkusen, Germany. • Simon Mahood, Wildlife Conservation Society, • Dr Nophea Sasaki, University of Hyogo, Japan. -

Fungus Gnats
Евразиатский энтомол. журнал 16(2): 119–128 © EUROASIAN ENTOMOLOGICAL JOURNAL, 2017 Fungus gnats (Diptera: Bolitophilidae, Diadocidiidae, Keroplatidae, Mycetophilidae) of the lower course of Anadyr River, Chukotskii Autonomnyi Okrug, Russia Ãðèáíûå êîìàðû (Diptera, Syrphidae) íèçîâèé ðåêè Àíàäûðü (×óêîòñêèé àâòîíîìíûé îêðóã, Ðîññèÿ) A.V. Polevoi*, A.V. Barkalov** À.Â. Ïîëåâîé*, À.Â. Áàðêàëîâ** * Forest Research Institute, Karelian Research Centre of the Russian Academy of Sciences, Pushkinskaya Str. 11, Petrozavodsk 185910 Russia. E-mail: [email protected]. * Институт леса КарНЦ РАН, ул. Пушкинская 11, Петрозаводск, 185910, Россия. E-mail: [email protected] ** Institute of Systematics and Ecology of Animals, Russian Academy of Sciences, Siberian Branch, Frunze Str. 11, Novosibirsk 630091 Russia. E-mail: [email protected]. ** Институт систематики и экологии животных СО РАН, ул. Фрунзе 11, Новосибирск 630091 Россия. Key words: fauna, fungus gnats, Anadyr River, Chukotka. Ключевые слова: фауна, грибные комары, река Анадырь, Чукотка. Abstract. The first data on the Fungus gnats fauna of Diadocidiidae, Ditomyiidae, Keroplatidae and Myceto- Chukotka are presented. 170 species belonging to the fami- philidae, belonging to the superfamily Sciaroidea (Diptera, lies Bolitophilidae, Diadocidiidae, Keroplatidae and Myce- Nematocera, Bibionomorpha). This is highly diverse group tophilidae were reported during two field seasons in 2013 with estimated number of species around 4500 in the and 2014 in the lower course of the Anadyr River. Eight world fauna and more than 1450 in Palaearctic [Søli et al., species are reported from Russia for the first time, two species are new for the Palaearctic and 27 species were 2000]. In the latter region, they seem to display an in- previously unknown in the eastern part of the Palaearctic; creasing diversity towards the North, with most species- 28 species are most probably undescribed taxa.