Impact of Processing Conditions on the Milk Clotting Activity of Crude Protease Extracted from Chinese Ginger
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
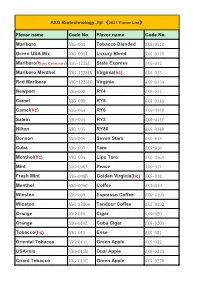
AXG Biotechnology.,Ltd (2021 Flavor List)
AXG Biotechnology.,ltd (2021 Flavor List) Flavor name Code No. Flavor name Code No. Marlboro AXG-001 Tobacco Blended AXG-011D Green USA Mix AXG-001A Luxury Blend AXG-011E Marlboro(highly commended) AXG-12281 State Express AXG-012 Marlboro Menthol AXG-12281M Virginia(hc) AXG-013 Red Marlboro AXG-12281R Virginia AXG-013A Newport AXG-002 RY4 AXG-014 Camel AXG-003 RY5 AXG-014A Camel(hc) AXG-06A RY6 AXG-014B Salem AXG-004 RY2 AXG-014C Hilton AXG-005 RY80 AXG-014D Benson AXG-006 Seven Stars AXG-015 Cuba AXG-007 Taro AXG-016 Menthol(hc) AXG-008 Lipu Taro AXG-016A Mint AXG-008A Peace AXG-017 Fresh Mint AXG-008B Golden Virginia(hc) AXG-018 Menthol AXG-008C Coffee AXG-019 Winston AXG-009 Espresso Coffee AXG-019A Winston AXG-33006 Tandoor Coffee AXG-019B Orange AXG-010 Cigar AXG-020 Orange AXG-010A Cuba Cigar AXG-020A Tobacco(hc) AXG-011 Esse AXG-021 Oriental Tobacco AXG-011A Green Apple AXG-022 USA-mix AXG-011B Dual Apple AXG-022A Crued Tobacco AXG-011C Green Apple AXG-022B Flavor name Code No. Flavor name Code No. Apple AXG-S0011 Vanilla AXG-040 Cigar Cherry AXG-023 Toffee AXG-040A Cherry AXG-023A Strawberry(hc) AXG-041 Cherry AXG-023B Milk Strawberry AXG-041A Cappuccino AXG-024 Tuscan AXG-042 Kent AXG-025 B&H AXG-043 Whisky AXG-026 Lark AXG-044 Cinnamon AXG-027 Lambert&Butler AXG-045 Parliament(hc) AXG-028 Gudang Garam AXG-046 Kool AXG-029 Surya Gudang Garam AXG-047 Dunhill AXG-030 Caster AXG-048 Have perfumed smoke AXG-031 Djarum Super AXG-049 Flue Cured AXG-031A Burley Tobacco AXG-050 Turkish Tobacco AXG-032 Blueberry AXG-051 French Pipe AXG-033 Blueberry AXG-051A Pipe Tobacco AXG-033A Old Captain AXG-052 Banana AXG-034 Captain Black Tobacco AXG-052A Banana AXG-034A Red Bull AXG-053 Deluxe 20035 Black Devil AXG-054 Davidoff AXG-036 Pineapple AXG-055 Cohiba AXG-037 Candy Hearts Pineapple AXG-055A Chocolate AXG-038 Almond AXG-056 Dark chocolate AXG-038A Toasted Almond AXG-056A Cream AXG-039 Plum AXG-057 Butterscotch AXG-039A Grape AXG-058 Flavor name Code No. -

STARTERS/BITES PANINI's PIZZAS MEZZE BOARDS DESSERT
Home Hotel Distillery Mixology Bar & Restaurant Events Store Knowledge Centre 0m Distillery Restaurant Opening Hours monday Closed tuesday Closed wednesday Closed Book Now thursday Closed friday Closed saturday Closed sunday Closed We're closed due to coronavirus outbreak. Bar & Restaurant Menus Prosecco Brunch Gin Masterclass Gin School Book Now Book Now Book Now BREAKFAST MENU Welsh Breakfast Pork and leek sausage, back bacon, hash brown, mushrooms, beans, grilled tomatoes and free range egg of your choice. Tea coffee and toast. £9.95 Vegetarian Breakfast Glamorgan vegan sausage, grilled tomato, hash brown, mushrooms, beans and free range egg of your choice. Tea, coffee and toast. £8.95 Vegan Breakfast Glamorgan vegan sausages, grilled tomatoes, mushrooms, baked beans and hash browns. Tea, coffee and toast. £8.95 Egg Benedict Toasted muffin with honey roasted ham, 2 soft poached eggs and our delicious hollandaise sauce. £8.95 Egg Royale Toasted muffin with salmon, 2 soft poached eggs and our delicious hollandaise sauce. £8.95 Omelette 3 egg omelette with a choice of ham, mushrooms, cheese, tomatoes and onions. £6.95 Porridge Served with a selection of fresh fruit and honey. £4.95 Pancake stack 4 thick American pancakes with syrup and fresh fruit. £5.95 STARTERS/BITES PIZZAS All of our pizzas are made with our own dough recipe and Marinara sauce made from Marzano tomatoes Goats cheese, carrot & orange salad (V) (GF) (NF) Roasted carrots with orange, goats cheese, with spinach and seeds, served Margherita (V) (VE) (NF) with fresh dressing. £6 Marinara sauce, fresh basil and buffalo mozzarella. £9 Quinoa Citrus Salad (V) (VE) (GF) (NF) Marinara (V) (VE) (DF) (NF) Quinoa marinated in orange and basil dressing with spinach, red onion, cucumber and red peppers. -

Microwave Cookbook 365
www.TheCookingMAP.com About 22 Authors 1. Healthy Land: https://www.thecookingmap.com/olivia-lopez 2. Bread Land: https://www.thecookingmap.com/emma-kim 3. Dessert Land: https://www.thecookingmap.com/sophia-garcia 4. Fruit and Vegetable Land: https://www.thecookingmap.com/emily-chan 5. Drink Land: https://www.thecookingmap.com/nathan-nelson 6. Pasta and Noodles Land: https://www.thecookingmap.com/jack-lemmon 7. Salad Land: https://www.thecookingmap.com/henry-fox 8. Appetizer and Snack Land: https://www.thecookingmap.com/ella-martinez 9. BBQ & Grilling Land: https://www.thecookingmap.com/ellie-lewis 10. Breakfast and Brunch Land: https://www.thecookingmap.com/anna-lee 11. Dinner Recipe Land: https://www.thecookingmap.com/victoria-lopez 12. {Everyday Cooking Land}: https://www.thecookingmap.com/sofia-rivera 13. Holiday Food Land: https://www.thecookingmap.com/chloe-webb 14. Cooking by Ingredient Land: https://www.thecookingmap.com/lily-li 15. Lunch Recipe Land : https://www.thecookingmap.com/lucy-liu 16. Main Dish Land: https://www.thecookingmap.com/benjamin-tee 17. Meat and Poultry Land: https://www.thecookingmap.com/nora-perry 18. Seafood Land: https://www.thecookingmap.com/mila-mason 19. Side Dish Land: https://www.thecookingmap.com/amelia-vega 20. Soup and Stew Land: https://www.thecookingmap.com/liam-fox 21. U.S.A Recipe Land: https://www.thecookingmap.com/lucas-neill 22. World Cuisine Land: https://www.thecookingmap.com/avery-moore Microwave Cookbook 365 Enjoy 365 Days with Amazing Microwave Recipes in Your Own Microwave Cookbook (Microwave Cookbook - Volume 1) Sofia Rivera Copyright: Published in the United States by Sofia Rivera / © SOFIA RIVERA Published on November 10, 2018 All rights reserved. -

Chengdu Kingdom
MAR/APR 2017 004 SKYInflight Magazine TIMES A Land of Phenomenal Florescence Living Like a Local in Chengdu Yang Mei Zhu Xie Street Beijing’s Most Artistic Hutong Kingdom of Wheaten Food A VILLAGE OF CRAFTMANSHIP CULINARY DELIGHT Providing the Promise of a Discover a Whole New World ‘Life Well Travelled’ in Seychelles Inflight magazine with the largest circulation around the world. SKY TIMES | Editor’s Letter A SPLENDID SPRINGTIME of BREATHTAKING BLOSSOMS o matter how harsh conditions get in winter, there is always something to look forward to — the arrival of spring. The energetic season brings blooming flowers, warmer weather and a Nmore relaxing lifestyle. In Chinese, there is an old saying: “Make your whole year’s plans in the spring, and your day’s plans early in the morning.” There is a very positive meaning behind the saying and at the start of the New Year of the Rooster, we encourage our readers to be adventurous and take advantage of the splendid springtime. In the March/April edition, Sky Times will embrace nature, showcase rural surroundings and present a plethora of breathtaking blossoms. To bring you a selection of spring scenery, we have chosen several spots, from Heqing, the village of craftsmanship and culinary delights in Yunnan Province (page 14), to Xinjiang, the northwestern border of China (page 40), and from the land of abundance in Chengdu (page 20), to Jining, the cradle of Confucius’ culture (page 24). In addition, we also take you to Africa to witness the stunning islands scattered across the eastern Indian Ocean and experience the sunshine, passion and hospitality of Seychelles (page 28). -
9781317510703.Pdf
Studies in Science Education in the Asia-Pacific Region Consistent with international trends, there is an active pursuit of more engaging sci- ence education in the Asia-Pacific region. The aim of this book is to bring together some examples of research being undertaken at a range of levels, from studies of cur- riculum and assessment tools, to classroom case studies, and investigations into mod- els of teacher professional learning and development. While neither a comprehensive nor definitive representation of the work that is being carried out in the region, the contributions – from China, Hong Kong, Taiwan, Korea, Japan, Singapore, Australia, and New Zealand – give a taste of some of the issues being explored, and the hopes that researchers have of positively influencing the types of science education experi- enced by school students. The purpose of this book is therefore to share contextual information related to science education in the Asia-Pacific region, as well as offering insights for conduct- ing studies in this region and outlining possible questions for further investigation. In addition, we anticipate that the specific resources and strategies introduced in this book will provide a useful reference for curriculum developers and science educators when they design school science curricula and science both pre-service and in-service teacher education programmes. The first section of the book examines features of science learners and learning, and includes studies investigating the processes associated with science conceptual learn- ing, scientific inquiry, model construction, and students’ attitudes towards science. The second section focuses on teachers and teaching. It discusses some more inno- vative teaching approaches adopted in the region, including the use of group work, inquiry-based instruction, developing scientific literacy, and the use of questions and analogies. -
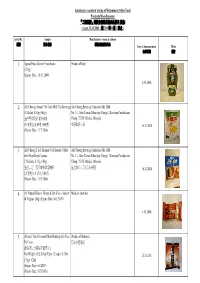
Type (7E) Miscellaneous 31.10.2008
Satisfactory results of testing of Melamine in Other Food Products(Miscellaneous) 「三聚氰胺」測試合格的其他食品樣本(其他) ( As at 31.10.2008 / 至 2008年10月31日止 ) Serial No. Sample Manufacturer’s name & address 編號 樣本名稱 製造商名稱及地址 Date of Announcement Photo 公布日期 相片 1 Agnesi Pesto Genove Pasta Sauce Product of Italy (185g) (Expiry Date : 18.11.2009) 8.10.2008 2 Aik Cheong Instant Teh Tarik Milk Tea Beverage Aik Cheong Beverage Industries Sdn. Bhd. 10 Sachet X 40g (400g ) No 1-1, Jalan Taman Teknologi Cheng 1, Kawasan Peridustrian 益昌香滑奶茶南洋風味 Cheng, 75250 Melaka, Malaysia 10 便利包 X 40克 (400克) 益昌飲品工業 16.10.2008 (Expiry Date : 11/7/2010) 3 Aik Cheong 2 in 1 Summer Café Instant Coffee Aik Cheong Beverage Industries Sdn. Bhd. with Non-Dairy Creamer No 1-1, Jalan Taman Teknologi Cheng 1, Kawasan Peridustrian 12 Sachets X 15g (180g) Cheng, 75250 Melaka, Malaysia 益昌二合一夏日咖啡即溶咖啡 益昌飲品工業 (馬來西亞) 16.10.2008 12 便利包 X 15克 (180克) (Expiry Date : 19/9/2009) 4 All Natural Bakery Honey & Oat Slice - Apricot Made in Australia & Yoghurt (80g) (Expiry Date: 16.6.2009) 4.10.2008 5 Almond Tofu Flavoured Mini Pudding with Nata Product of Malaysia De Coco 馬來西亞製造 迷你杏仁豆腐花味椰果布丁 Net Weight /凈重:180g (15g x 12 cups) /6.35oz 20.10.2008 (15g x 12個) (Expiry Date: 6/5/2009) (Expiry Date: 15/5/2009) Satisfactory results of testing of Melamine in Other Food Products(Miscellaneous) 「三聚氰胺」測試合格的其他食品樣本(其他) ( As at 31.10.2008 / 至 2008年10月31日止 ) Serial No. Sample Manufacturer’s name & address 編號 樣本名稱 製造商名稱及地址 Date of Announcement Photo 公布日期 相片 6 Alpen Swiss Style Muesli (560g)(Expiry Date: Manufacturer: WEETABIX 19.5.2009) Address: Weetabix Ltd. -

Cooking With
COOKING WITH 5000 YEARS OF PERUVIAN CUISINE 5000 YEARS OF PERUVIAN CUISINE COOKING WITH PERUVIAN AVOCADO COMMISSION Enrique Camet Chairman Xavier Fco. Equihua President & CEO 717 D Street, NW Suite 310 Washington, D.C. 20004 phone (202) 626-0560 fax (202) 393-5728 [email protected] www.avocadosfromperu.com PROHASS Jimmy Bosworth Chairman Arturo Medina Managing Director Partner Association in Peru Nicolás Arriola 314 Of.1101 La Victoria, Lima Peru Phone (511) 225-1626 [email protected] www.prohass.com.pe PRESENTATION..........................................................................4 Foreword....................................................................................8 Introduction..............................................................................10 Unveiling the secret of Peru........................................................12 The Summer Avocado .................................................................14 Tips & selection..........................................................................16 Step by step...............................................................................18 Eat healthy, not heavy................................................................20 Ingredients in Peruvian cuisine....................................................24 Historical chapters......................................................................32 Part 1 ........................................................................................24 Part 2........................................................................................44 -

HCS Product List - by Brand Name
HCS Product List - by Brand Name Brand and Product Name Package Size (Weight) (Lim Traders) Chicken Breaded Patties 1.8kg 100PLUS ACTIVE Non-Carbonated Drink 1.5L 100PLUS ACTIVE Non-Carbonated Drink 12 x 300ml 100PLUS ACTIVE Non-Carbonated Drink 300ml 100PLUS ACTIVE Non-Carbonated Drink 500ml 100PLUS ACTIVE Non-Carbonated Drink 6 x 300ml 100PLUS ACTIVE NON-CARBONATED ISOTONIC DRINK 1.5L 100PLUS ACTIVE NON-CARBONATED ISOTONIC DRINK 300ml 100PLUS ACTIVE NON-CARBONATED ISOTONIC DRINK 500ml 100PLUS Blackcurrant 1.5L 100PLUS Blackcurrant 12 x 1.5L 100PLUS Blackcurrant 24 x 500ml 100PLUS Blackcurrant 500ml 100PLUS EDGE NON-CARBONATED ISOTONIC DRINK 1.5L 100PLUS EDGE NON-CARBONATED ISOTONIC DRINK 300ml 100PLUS EDGE NON-CARBONATED ISOTONIC DRINK 500ml 100PLUS Lemon Lime 1.5L 100PLUS Lemon Lime 325ml 100PLUS Lemon Lime 500ml 100PLUS Orange 1.5L 100PLUS Orange 500ml 100PLUS Original 1.5L 100PLUS Original 12 x 1.5L 100PLUS Original 12 x 325ml 100PLUS Original 24 x 325ml 100PLUS Original 24 x 500ml 100PLUS Original 325ml 100PLUS Original 500ml 100PLUS Original 6 x 325ml Brand and Product Name Package Size (Weight) 100PLUS ORIGINAL ISOTONIC DRINK 1.5L 100PLUS ORIGINAL ISOTONIC DRINK 325ml 100PLUS ORIGINAL ISOTONIC DRINK 500ml 100PLUS Zero Sugar 1.5L 100PLUS Zero Sugar 12 x 1.5L 100PLUS Zero Sugar 24 x 325ml 100PLUS Zero Sugar 24 x 500ml 100PLUS Zero Sugar 325ml 100PLUS Zero Sugar 4 x 6 x 325ml 100PLUS Zero Sugar 500ml 100PLUS Zero Sugar 6 x 325ml 333 Super Refined Blended Vegetable Oil 17kg 3A 100% Pure Black Sesame Oil 320ml 3A 100% Pure Black Sesame -

GOOD FOOD IS a JOURNEY BEST SHARED with FAMILY & FRIENDS Niyama Private Islands Is Home to Tribal Restaurant, the First and Only Afro Latin Fusion in the Maldives
GOOD FOOD IS A JOURNEY BEST SHARED WITH FAMILY & FRIENDS Niyama Private Islands is home to Tribal restaurant, the first and only Afro Latin fusion in the Maldives. The menu is a showcase of dishes that reflects a celebration of rich culinary heritage, embracing the ancient spice routes and the diversity of cultures that have influenced Africa and South and Central American Tribal cuisines. House specialties includes the abundant fresh produce found along the shores and in the waters of the two continents, African Game and artisanal meat cuts. An authentic Boma or Cauchu, live and interactive style of dining. Tribal provides a uniquely authentic cultural experience that showers the sense with the tastes, sight, sounds, touch and smell of traditional tribal cooking. We wish you a memorable experience with us and trust that your culinary adventure will capture your imagination and inspire your senses. Let your journey begins…………… Tribal Afro Latin Cuisine Camp Fire Starter 225 for two Starter Octopus and Snapper Numus (R,SP) Shaved Octopus, white snapper sashimi, coconut, habanero, lime Main Course Tribal Tasting Platter Mahi-Mahi fish fillet, harrisa prawns, Karoo lamb cutlet, baby chicken, Braai vegetables, chimichurri, biltong sauces Dessert Valrhona Chocolate Story Guanaja, Caraibe and Jivara mousse chocolate soil, chocolate comb, lemon gel Reyneke Organic Sauvignon Blanc Semillon, Stellenbosch, South Africa 2017 Pinotage-Cabernet Sauvignon Kanonkop Kadette, Stellenbosch 2016 Kindly Notify One of Our Team Members if You Have Any Allergic Intolerance P = Pork, R = Raw, V= Vegetarian N = contains nuts, SP = Spicy Food ɸ =Symbol is a Must Try Tribal Signature Dish All Prices are in U.S. -

Culinary Travel in Peru the Best Food and Drink and Where to Find It
Culinary Travel In Peru The Best Food and Drink and Where to Find It Foreword by Pedro Miguel Schiaffino Culinary Travel in Peru The Best Food and Drink and Where to Find It Foreword by Pedro Miguel Schiaffino Copyright © 2015 Aracari Travel Jr. Schell 237 # 602 - MIRAFLORES - LIMA – PERU T: +511 651 2424 Layout & design by Simon Ross-Gill Front cover photograph courtesy of ámaZ restaurant. Culinary Travel in Peru: The Best Food and Drink and Where to Find It Contents Foreword ........................................................................................................Pages 6-7 Preface ............................................................................................................Pages 8-9 Regional Styles ..............................................................................................Pages 10-11 Dishes to Try ...................................................................................................Pages 12-15 Fun Food Facts ...............................................................................................Pages 16-17 Need to know ................................................................................................Pages 18-19 Lima Introduction ..............................................................................................Pages 22-23 Culinary Experiences ..................................................................................Pages 24-27 Listings .......................................................................................................Pages -

Soup Cold Selection
Soup Vegetarian Sopa de Zapallo Peruvian pumpkin soup Non -Vegetarian Caldo de Albondigas Mexican meat ball soup Cold Selection Vegetarian Zucchini Tiradito Grilled zucchini, olive mayonnaise, crunchy quinoa, goat cheese Causa Rellena Stuffed potato salad Truffled Watermelon Salad Watermelon, roasted eggplant, feta cheese and balsamic vinaigrette Glass Noodle Salad Non- Vegetarian Tuna Tiradito Tuna,chia seeds,yellow Aji paste Chicken Causa Chicken, potatoes, Huancaina sauce Lima Ceviche Indian seabass, Red onion, Leche de tigre Mini Cubano Ham and cheese sandwich Hot Selection Vegetarian Yucca Frita Fried yucca chips Vegetable Quesadillas Bell pepper, zucchini, habanero chili, cottage cheese and Feta Croquettes Bread crumbed Uruguayan potato and cheese croquettes Tinga Mushroom Tostadas Mushroom, tinga sauce, sour cream Non -Vegetarian Chorizo Molettes Spanish chorizo, frijoles, salsa backed with cheese on a baguette Puerto Rican Chillo Fritto Fried red snapper and plantain tostones Chifa Chicken Chifa style chicken stir fry with scallions and ginger Lamb Taco Pulled lamb leg with anticuchera sauce and salsa criolla Churrasco Grill Vegetarian Smoke Grilled Yucca Grilled yucca, green tomato salsa and salsa criolla Corn on the Cob Grilled corn, chipotle rub, cilantro sour cream Grilled Miso Aubergine Miso and honey glazed baby eggplant Non - Vegetarian Choripan Grilled Argentinian chorizo in a baguette Chimichurri Chicken Liver Chicken liver and Andean herb chimichurri Gochujang Chicken Chicken thigh, Gochujang chili paste and honey Main -

Oscars® Governors Ball Menu Sunday, March 4, 2018
Oscars® Governors Ball Menu Sunday, March 4, 2018 Menu created by Wolfgang Puck and the Wolfgang Puck Catering Team, including VP of Culinary Eric Klein, Chef Connor Shanahan, and pastry team Kamel Guechida, Monica Ng, Garry Larduinat and Jason Lemonnier Amuse Bouche Parmesan Bread Sticks Everything Bagel Pretzel Bite, Scamorza Cream Cheese, Tomato Confit Aged Vermont Cheddar Cheese Aged Parmesan Cheese Candied Spiced Mixed Nuts, Pineapple Dried Fruit Apricot Macadamia Nut Lavosh Passed Hors d’Oeuvre Edamame and Black Truffle Pot Stickers (V) Miyazaki Wagyu Beef Tartare, Puffed Black Rice, Togarashi * Of the 28,000 Japanese Black cattle handled by JA Miyazaki, only 18,000 meet the criteria to become Miyazaki Wagyu. Tiny Taro Taco, Spiced Eggplant, Lime Pickle (V) Smoked Salmon Oscars Miniature Wagyu Burger with Sharp Cheddar and Remoulade Signature Pizzas Spicy Tuna Tartare in Sesame Miso Cones Crab Stuffed Hibiscus, Lemon, Pomegranate Explosion of Rhubarb, Thai Flavors (V) Small Plates Crispy Brussels Sprouts, Pea Shoots, Mezze Flavors (V) Frozen Beets, Ginger Milk Curd, Yuzu, Ice Lettuce Asparagus, Shiro Miso, Watercress, Cherry Tomatoes, Sancho Pepper (V) White Asparagus Soup, Caviar, Yukon Potato Croutons Potato Caviar 2.0 Black Truffle Chicken Pot Pie Winter Truffle Baked Cavatappi and Cheese Mini Pea and Carrot Ravioli, Black Truffle, Cipollini Onions Miyazaki Wagyu Beef, Forbidden Black Rice, Crunchy Red Beets, Char Siu Cabbage Black Bass, Eggplant, Salsa Verde, Teardrop Peppers Housemade Spinach Campanelle, English Peas, Cipollini