Herpetologists' League
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
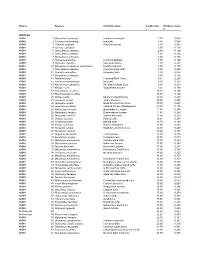
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

Kinosternon Subrubrum (Bonnaterre 1789) – Eastern Mud Turtle
Conservation Biology of Freshwater Turtles and Tortoises: A Compilation ProjectKinosternidae of the IUCN SSC — KinosternonTortoise and Freshwater subrubrum Turtle Specialist Group 101.1 A.G.J. Rhodin, J.B. Iverson, P.P. van Dijk, K.A. Buhlmann, P.C.H. Pritchard, and R.A. Mittermeier, Eds. Chelonian Research Monographs (ISSN 1088-7105) No. 5, doi:10.3854/crm.5.101.subrubrum.v1.2017 © 2017 by Chelonian Research Foundation and Turtle Conservancy • Published 17 September 2017 Kinosternon subrubrum (Bonnaterre 1789) – Eastern Mud Turtle WALTER E. MESHAKA, JR.1, J. WHITFIELD GIBBONS2, DANIEL F. HUGHES3, MICHAEL W. KLEMENS4, AND JOHN B. IVERSON5 1State Museum of Pennsylvania, 300 North Street, Harrisburg, Pennsylvania 17120 USA [[email protected]]; 2Savannah River Ecology Lab, Drawer E, Aiken, South Carolina 29802 USA [[email protected]]; 3University of Texas at El Paso, El Paso, Texas 79968 USA [[email protected]]; 4Department of Herpetology, American Museum of Natural History, Central Park West at 79th Street, New York, New York 10024 USA [[email protected]]; 5Department of Biology, Earlham College, Richmond, Indiana 47374 USA [[email protected]] SUMMARY. — The Eastern Mud Turtle, Kinosternon subrubrum (Family Kinosternidae), is a small (carapace length 85 to 120 mm) polytypic species of the eastern and central United States. All three historically recognized subspecies (K. s. subrubrum, K. s. steindachneri, and K. s. hippocrepis) are semi-aquatic turtles that inhabit much of the U.S. Atlantic and Gulf Coastal Plains. The Florida taxon (K. s. steindachneri) appears to represent a distinct species, but we continue to treat it as a subspecies for the purposes of this account. -

Standard Common and Current Scientific Names for North American Amphibians, Turtles, Reptiles & Crocodilians
STANDARD COMMON AND CURRENT SCIENTIFIC NAMES FOR NORTH AMERICAN AMPHIBIANS, TURTLES, REPTILES & CROCODILIANS Sixth Edition Joseph T. Collins TraVis W. TAGGart The Center for North American Herpetology THE CEN T ER FOR NOR T H AMERI ca N HERPE T OLOGY www.cnah.org Joseph T. Collins, Director The Center for North American Herpetology 1502 Medinah Circle Lawrence, Kansas 66047 (785) 393-4757 Single copies of this publication are available gratis from The Center for North American Herpetology, 1502 Medinah Circle, Lawrence, Kansas 66047 USA; within the United States and Canada, please send a self-addressed 7x10-inch manila envelope with sufficient U.S. first class postage affixed for four ounces. Individuals outside the United States and Canada should contact CNAH via email before requesting a copy. A list of previous editions of this title is printed on the inside back cover. THE CEN T ER FOR NOR T H AMERI ca N HERPE T OLOGY BO A RD OF DIRE ct ORS Joseph T. Collins Suzanne L. Collins Kansas Biological Survey The Center for The University of Kansas North American Herpetology 2021 Constant Avenue 1502 Medinah Circle Lawrence, Kansas 66047 Lawrence, Kansas 66047 Kelly J. Irwin James L. Knight Arkansas Game & Fish South Carolina Commission State Museum 915 East Sevier Street P. O. Box 100107 Benton, Arkansas 72015 Columbia, South Carolina 29202 Walter E. Meshaka, Jr. Robert Powell Section of Zoology Department of Biology State Museum of Pennsylvania Avila University 300 North Street 11901 Wornall Road Harrisburg, Pennsylvania 17120 Kansas City, Missouri 64145 Travis W. Taggart Sternberg Museum of Natural History Fort Hays State University 3000 Sternberg Drive Hays, Kansas 67601 Front cover images of an Eastern Collared Lizard (Crotaphytus collaris) and Cajun Chorus Frog (Pseudacris fouquettei) by Suzanne L. -

Proposed Amendment to 21CFR124021
Richard Fife 8195 S. Valley Vista Drive Hereford, AZ 85615 December 07, 2015 Division of Dockets Management Food and Drug Administration 5630 Fishers Lane, rm. 1061 Rockville, MD 20852 Reference: Docket Number FDA-2013-S-0610 Proposed Amendment to Code of Federal Regulations Title 21, Volume 8 Revised as of April 1, 2015 21CFR Sec.1240.62 Dear Dr. Stephen Ostroff, M.D., Acting Commissioner: Per discussion with the Division of Dockets Management staff on November 10, 2015 Environmental and Economic impact statements are not required for petitions submitted under 21CFR Sec.1240.62 CITIZEN PETITION December 07, 2015 ACTION REQUESTED: I propose an amendment to 21CFR Sec.1240.62 (see exhibit 1) as allowed by Section (d) Petitions as follows: Amend section (c) Exceptions. The provisions of this section are not applicable to: By adding the following two (2) exceptions: (5) The sale, holding for sale, and distribution of live turtles and viable turtle eggs, which are sold for a retail value of $75 or more (not to include any additional turtle related apparatuses, supplies, cages, food, or other turtle related paraphernalia). This dollar amount should be reviewed every 5 years or more often, as deemed necessary by the department in order to make adjustments for inflation using the US Department of Labor, Bureau of labor Statistics, Consumer Price Index. (6) The sale, holding for sale, and distribution of live turtles and viable turtle eggs, which are listed by the International Union for Conservation of Nature and Natural Resources (IUCN) Red List as Extinct In Wild, Critically Endangered, Endangered, or Vulnerable (IUCN threatened categorizes). -

Turtles of the World, 2010 Update: Annotated Checklist of Taxonomy, Synonymy, Distribution, and Conservation Status
Conservation Biology of Freshwater Turtles and Tortoises: A Compilation ProjectTurtles of the IUCN/SSC of the World Tortoise – 2010and Freshwater Checklist Turtle Specialist Group 000.85 A.G.J. Rhodin, P.C.H. Pritchard, P.P. van Dijk, R.A. Saumure, K.A. Buhlmann, J.B. Iverson, and R.A. Mittermeier, Eds. Chelonian Research Monographs (ISSN 1088-7105) No. 5, doi:10.3854/crm.5.000.checklist.v3.2010 © 2010 by Chelonian Research Foundation • Published 14 December 2010 Turtles of the World, 2010 Update: Annotated Checklist of Taxonomy, Synonymy, Distribution, and Conservation Status TUR T LE TAXONOMY WORKING GROUP * *Authorship of this article is by this working group of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, which for the purposes of this document consisted of the following contributors: ANDERS G.J. RHODIN 1, PE T ER PAUL VAN DI J K 2, JOHN B. IVERSON 3, AND H. BRADLEY SHAFFER 4 1Chair, IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Chelonian Research Foundation, 168 Goodrich St., Lunenburg, Massachusetts 01462 USA [[email protected]]; 2Deputy Chair, IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Conservation International, 2011 Crystal Drive, Suite 500, Arlington, Virginia 22202 USA [[email protected]]; 3Department of Biology, Earlham College, Richmond, Indiana 47374 USA [[email protected]]; 4Department of Evolution and Ecology, University of California, Davis, California 95616 USA [[email protected]] AB S T RAC T . – This is our fourth annual compilation of an annotated checklist of all recognized and named taxa of the world’s modern chelonian fauna, documenting recent changes and controversies in nomenclature, and including all primary synonyms, updated from our previous three checklists (Turtle Taxonomy Working Group [2007b, 2009], Rhodin et al. -

Minnesota Herpetological Society
The newsletter of the Minnesota Herpetological Society January Meeting Notice General Meeting will be January 3rd, 2014 January Speaker Jeff LeClere—Radio Telemetry of Bullsnakes and Plains Hog-nosed Snakes in Minnesota January 2014 Volume 34 Number 1 BOARD OF DIRECTORS President Chris Smith 612.275.9737 [email protected] C/O Bell Museum of Natural History Vice President 10 Church Street Southeast Peter Tornquist 952.797.6515 Minneapolis, Minnesota, 55455-0104 [email protected] Stay informed! Join us on our forums! Recording Secretary And, you can still leave us a Voice Mail: 612.326.6516 Membership Secretary The purpose of the Minnesota Herpetological Society is to: Micole Hendricks 651.356.1669 [email protected] • Further the education of the membership and the general public in care and captive propagation of reptiles and amphibians; Treasurer • Educate the members and the general public in the ecological role of Nancy Haig 763.434.8684 reptiles and amphibians; [email protected] • Promote the study and conservation of reptiles and amphibians. Newsletter Editor The Minnesota Herpetological Society is a non-profit, tax-exempt organiza- Ellen Heck 763.593.5414 tion. Membership is open to all individuals with an interest in amphibians [email protected] and reptiles. The Minnesota Herpetological Society Newsletter is published monthly to provide its members with information concerning the society’s Members at Large activities and a media for exchanging information, opinions and resources. Heather Clayton 612.886.7175 [email protected] General Meetings are held at Borlaug Hall, Room 335 on the St. Paul Cam- pus of the University of Minnesota, on the first Friday of each month (unless Rebecca Markowitz 409.750.0235 there is a holiday conflict). -

Global Turtle Demand and Illegal Trafficking
Global Turtle Demand and Illegal Trafficking Staff Report February 20, 2020 The purpose of this presentation is to provide the Commission with an update on the global demand on turtles and actions we are taking to combat illegal trafficking here in Florida. Division: Division of Law Enforcement Authors: Colonel Curtis Brown Report Date: February 20, 2020 Photo Credit: FWC Photo (Pierson Hill) Ornate diamondback terrapin (Malaclemys terrapin macrospilota) Turtle Life Cycle ■ Turtles exhibit a life history characterized by slow growth and late maturity. ■ Wild turtle populations sustain their greatest natural loss in the egg and juvenile stages, but experience very low natural adult mortality. ■ Once depleted, a turtle population recovers very slowly (several decades), if at all. Photo Credit: FWC Photo (Jonathan Mays) Keys mud turtles (Kinosternon baurii) Current Status of Turtles in Florida Florida is home to approximately 33 species of turtles and tortoises. ■ 23 Freshwater species ■ 1 Terrestrial species ■ 6 saltwater species ■ 3 nonnative species The reason we say approximately 33 species is because views on species taxonomy may differ. Some species may still be under review for subspecies classification. 23 freshwater species: Southeastern mud turtle) • common snapping turtle • Eastern loggerhead musk turtle • Suwannee alligator snapping turtle • Intermediate Musk Turtle (aka • alligator snapping turtle Aliflora musk turtle) • spotted turtle • Eastern musk turtle (aka stinkpot) • chicken turtle • Florida softshell • Barbour’s map -

The Stratigraphic Position of Fossil Vertebrates from the Pojoaque Member of the Tesuque Formation (Middle Miocene, Late Barstovian) Near Española, New Mexico
Stephen F. Austin State University SFA ScholarWorks Electronic Theses and Dissertations 5-9-2016 THE STRATIGRAPHIC POSITION OF FOSSIL VERTEBRATES FROM THE POJOAQUE MEMBER OF THE TESUQUE FORMATION (MIDDLE MIOCENE, LATE BARSTOVIAN) NEAR ESPAÑOLA, NEW MEXICO Garrett R. Williamson Stephen F. Austin State University, [email protected] Follow this and additional works at: https://scholarworks.sfasu.edu/etds Part of the Geochemistry Commons, Geology Commons, Geomorphology Commons, Paleobiology Commons, Paleontology Commons, and the Stratigraphy Commons Tell us how this article helped you. Repository Citation Williamson, Garrett R., "THE STRATIGRAPHIC POSITION OF FOSSIL VERTEBRATES FROM THE POJOAQUE MEMBER OF THE TESUQUE FORMATION (MIDDLE MIOCENE, LATE BARSTOVIAN) NEAR ESPAÑOLA, NEW MEXICO" (2016). Electronic Theses and Dissertations. 41. https://scholarworks.sfasu.edu/etds/41 This Thesis is brought to you for free and open access by SFA ScholarWorks. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of SFA ScholarWorks. For more information, please contact [email protected]. THE STRATIGRAPHIC POSITION OF FOSSIL VERTEBRATES FROM THE POJOAQUE MEMBER OF THE TESUQUE FORMATION (MIDDLE MIOCENE, LATE BARSTOVIAN) NEAR ESPAÑOLA, NEW MEXICO Creative Commons License This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 4.0 License. This thesis is available at SFA ScholarWorks: https://scholarworks.sfasu.edu/etds/41 THE STRATIGRAPHIC POSITION OF FOSSIL VERTEBRATES FROM THE POJOAQUE MEMBER OF THE TESUQUE FORMATION (MIDDLE MIOCENE, LATE BARSTOVIAN) NEAR ESPAÑOLA, NEW MEXICO By GARRETT ROSS WILLIAMSON, B.S. Presented to the Faculty of the Graduate School of Stephen F. Austin State University In Partial Fulfillment Of the Requirements For the Degree of Master of Science STEPHEN F. -

Journal of Vertebrate Paleontology Supplementary
JOURNAL OF VERTEBRATE PALEONTOLOGY SUPPLEMENTARY DATA Fossil musk turtles (Kinosternidae, Sternotherus) from the late Miocene–early Pliocene (Hemphillian) of Tennessee and Florida JASON R. BOURQUE*, 1 and BLAINE W. SCHUBERT2 1Division of Vertebrate Paleontology, Florida Museum of Natural History, University of Florida, Gainesville, Florida 32611, U.S.A., [email protected]; 2Don Sundquist Center of Excellence in Paleontology and Department of Geosciences, East Tennessee State University, Johnson City, Tennessee 37614, U.S.A., [email protected] *Corresponding author TABLES TABLE S1. Updated vertebrate fauna from the Gray Fossil Site. Compiled from Wallace and Wang (2004), Bentley et al. (2008), Hulbert et al. (2009), Boardman and Schubert (2011), Schubert and Mead (2011), Mead et al. (2012). Additional unpublished identifications by S. C. Wallace, J. I. Mead, B.W.S., and J.R.B.. Osteichthyes Centrarchidae Amphibia Anura Ranidae Rana sp. Bufonidae Hylidae Caudata Ambystomatidae Ambystoma sp. Plethodontidae Plethodon sp. Desmognathus sp. Salamandridae Notophthalmus sp. Reptilia Testudines Chelydridae Chelydra sp. Kinosternidae Sternotherus palaeodorus, n. sp. Emydidae Chrysemys sp. Trachemys sp. aff. Emydoidea or Emys sp. Terrapene sp. Testudinidae Hesperotestudo sp. Crocodylia Alligatoridae Alligator sp. Squamata Anguidae Ophisaurinae Scincidae Helodermatidae TABLE 1S. (Continued) Heloderma sp. Colubridae Natricinae Colubrinae Viperidae Aves Anatidae Aix sp. Anas spp. Odontophoridae Rallidae Scolopacidae Scolopax sp. Tytonidae Mammalia Xenarthra Megalonychidae Insectivora Soricidae minimum 3 spp. present Talpidae minimum 2 spp. present Chiroptera Lagomorpha Leporidae Rodentia minimum 6 spp. present Proboscidea Gomphotheriidae Perissodactyla Equidae Cormohipparion emsliei Tapiridae Tapirus polkensis Rhinocerotidae Teleoceras sp. Artiodactyla Tayassuidae Prosthennops sp. sp. indet. Camelidae cf. Megatylopus sp. Carnivora TABLE S1. (Continued) Felidae cf. Machairodus sp. -

Billing Code 4333–15 DEPARTMENT of THE
This document is scheduled to be published in the Federal Register on 04/21/2021 and available online at Billing Code 4333–15 federalregister.gov/d/2021-07402, and on govinfo.gov DEPARTMENT OF THE INTERIOR Fish and Wildlife Service 50 CFR Part 17 [Docket No. FWS–R8–ES–2013–0011; FF09E21000 FXES11110900000 212] RIN 1018–BE29 Endangered and Threatened Wildlife and Plants; Designation of Critical Habitat for the Western Distinct Population Segment of the Yellow-billed Cuckoo AGENCY: Fish and Wildlife Service, Interior. ACTION: Final rule. SUMMARY: We, the U.S. Fish and Wildlife Service (Service), designate critical habitat for the western distinct population segment of the yellow-billed cuckoo (western yellow- billed cuckoo) (Coccyzus americanus) under the Endangered Species Act. In total, approximately 298,845 acres (120,939 hectares) are now being designated as critical habitat in Arizona, California, Colorado, Idaho, New Mexico, Texas, and Utah. This rule extends the Act’s protections to critical habitat for this species. DATES: This rule is effective [INSERT DATE 30 DAYS AFTER DATE OF PUBLICATION IN THE FEDERAL REGISTER]. ADDRESSES: This final rule is available on the Internet at http://www.regulations.gov, and the Sacramento Fish and Wildlife Office website at http://www.fws.gov/sacramento. Comments and materials we received, as well as supporting documentation we used or developed in preparing this rule, are available for public inspection at http://www.regulations.gov at Docket No. FWS–R8–ES–2013–0011. The coordinates or plot points or both from which the maps are generated are included in the decisional record for this critical habitat designation and are available at http://www.regulations.gov at Docket No. -

Scientific and Standard English Names of Amphibians and Reptiles of North America North of Mexico, with Comments Regarding Confidence in Our Understanding
SCIENTIFIC AND STANDARD ENGLISH NAMES OF AMPHIBIANS AND REPTILES OF NORTH AMERICA NORTH OF MEXICO, WITH COMMENTS REGARDING CONFIDENCE IN OUR UNDERSTANDING SEVENTH EDITION COMMITTEE ON STANDARD ENGLISH AND SCIENTIFIC NAMES BRIAN I. CROTHER (Committee Chair) STANDARD ENGLISH AND SCIENTIFIC NAMES COMMITTEE Jeff Boundy, Frank T. Burbrink, Jonathan A. Campbell, Brian I. Crother, Kevin de Queiroz, Darrel R. Frost, David M. Green, Richard Highton, John B. Iverson, Fred Kraus, Roy W. McDiarmid, Joseph R. Mendelson III, Peter A. Meylan, R. Alexander Pyron, Tod W. Reeder, Michael E. Seidel, Stephen G. Tilley, David B. Wake Official Names List of American Society of Ichthyologists and Herpetologists Canadian Association of Herpetology Canadian Amphibian and Reptile Conservation Network Partners in Amphibian and Reptile Conservation Society for the Study of Amphibians and Reptiles The Herpetologists’ League 2012 SOCIETY FOR THE STUDY OF AMPHIBIANS AND REPTILES HERPETOLOGICAL CIRCULAR NO. 39 Published August 2012 © 2012 Society for the Study of Amphibians and Reptiles John J. Moriarty, Editor 3261 Victoria Street Shoreview, MN 55126 USA [email protected] Single copies of this circular are available from the Publications Secretary, Breck Bartholomew, P.O. Box 58517, Salt Lake City, Utah 84158–0517, USA. Telephone and fax: (801) 562-2660. E-mail: [email protected]. A list of other Society publications, including Facsimile Reprints in Herpetology, Herpetologi- cal Conservation, Contributions to Herpetology, and the Catalogue of American Amphibians and Reptiles, will be sent on request or can be found at the end of this circular. Membership in the Society for the Study of Amphibians and Reptiles includes voting privledges and subscription to the Society’s technical Journal of Herpe- tology and news-journal Herpetological Review, both are published four times per year. -

Single Page Version
Herpetology in Bogey Creek Florida is home to a wide variety of unique reptiles and amphibians, some of which call Bogey Creek Preserve their home. These trails pass through several habitats which are prime for different species of animals. These animals, colloquially referred to as “herps” (as in herpetology), take full advantage of each of them. They often go unnoticed and unappreciated, but they serve important roles in their environment and are very diverse indeed. To learn more about notable “herps” that inhabit the area and how to identify them, take a search through NFLT’s Bogey Creek Herpetology Field Guide! WELCOME Be sure to take photos of any species you find at Bogey Creek Preserve and tag #bogeycreekpreserve. NORTH FLORIDA LAND TRUST | 1 BARKING TREE FROG Hyla gratiosa FWC photo by Kevin Enge METAMORPH Photo by John P. Friel DESCRIPTION Barking treefrogs are quite large and range from 2-3 in (5-7 cm). In fact, they are the largest species of treefrog in the southeastern corner of the country and among the largest species in the United States. They are known for their large toepads. Coloration varies radically, although they generally have earth-tone pigments. They can change the color of their skin in a similar manner to chameleons. They may be a lime green color, dark brown, gray, or even have a slight purplish hue. Large and bold spots adorn their backs and legs with dark borders, and gold dots may be scattered across the body, but these patterns usually are faded when they are bright green.