Intracellular Trafficking of HBV Particles
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Formation of Virus-Like Particles from Human Cell Lines Exclusively Expressing Influenza Neuraminidase
Journal of General Virology (2010), 91, 2322–2330 DOI 10.1099/vir.0.019935-0 Formation of virus-like particles from human cell lines exclusively expressing influenza neuraminidase Jimmy C. C. Lai,1 Wallace W. L. Chan,1 Franc¸ois Kien,1 John M. Nicholls,2 J. S. Malik Peiris1,3 and Jean-Michel Garcia1 Correspondence 1HKU-Pasteur Research Centre, Hong Kong SAR Jean-Michel Garcia 2Department of Pathology, The University of Hong Kong, Hong Kong SAR [email protected] 3Department of Microbiology, The University of Hong Kong, Hong Kong SAR The minimal virus requirements for the generation of influenza virus-like particle (VLP) assembly and budding were reassessed. Using neuraminidase (NA) from the H5N1 and H1N1 subtypes, it was found that the expression of NA alone was sufficient to generate and release VLPs. Biochemical and functional characterization of the NA-containing VLPs demonstrated that they were morphologically similar to influenza virions. The NA oligomerization was comparable to that of the live virus, and the enzymic activity, whilst not required for the release of NA-VLPs, was Received 8 January 2010 preserved. Together, these findings indicate that NA plays a key role in virus budding and Accepted 23 May 2010 morphogenesis, and demonstrate that NA-VLPs represent a useful tool in influenza research. INTRODUCTION membrane. HA binds to sialic acid receptors on the cell surface and mediates the fusion process (Matrosovich et al., Influenza A viruses are lipid-enveloped members of the 2006), whereas NA cleaves the terminal sialic acids from family Orthomyxoviridae. They contain eight negative-sense, the cell-surface glycans to facilitate release of the progeny single-stranded RNA segments encoding ten viral proteins. -

Discovery of a Highly Divergent Hepadnavirus in Shrews from China
Virology 531 (2019) 162–170 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/virology Discovery of a highly divergent hepadnavirus in shrews from China T Fang-Yuan Niea,b,1, Jun-Hua Tianc,1, Xian-Dan Lind,1, Bin Yuc, Jian-Guang Xinge, Jian-Hai Caof, ⁎ ⁎⁎ Edward C. Holmesg,h, Runlin Z. Maa, , Yong-Zhen Zhangb,h, a State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China b State Key Laboratory for Infectious Disease Prevention and Control; Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases; Department of Zoonoses, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing, China c Wuhan Center for Disease Control and Prevention, Wuhan, China d Wenzhou Center for Disease Control and Prevention, Wenzhou, Zhejiang Province, China e Wencheng Center for Disease Control and Prevention, Wencheng, Zhejiang Province, China f Longwan Center for Disease Control and Prevention, Longwan District, Wenzhou, Zhejiang Province, China g Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life and Environmental Sciences and Sydney Medical School, The University of Sydney, Sydney, NSW 2006, Australia h Shanghai Public Health Clinical Center & Institute of Biomedical Sciences, Fudan University, Shanghai, China ARTICLE INFO ABSTRACT Keywords: Limited sampling means that relatively little is known about the diversity and evolutionary history of mam- Hepadnaviruses malian members of the Hepadnaviridae (genus Orthohepadnavirus). An important case in point are shrews, the Shrews fourth largest group of mammals, but for which there is limited knowledge on the role they play in viral evo- Phylogeny lution and emergence. -

A SARS-Cov-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing
A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing Supplementary Information Supplementary Discussion All SARS-CoV-2 protein and gene functions described in the subnetwork appendices, including the text below and the text found in the individual bait subnetworks, are based on the functions of homologous genes from other coronavirus species. These are mainly from SARS-CoV and MERS-CoV, but when available and applicable other related viruses were used to provide insight into function. The SARS-CoV-2 proteins and genes listed here were designed and researched based on the gene alignments provided by Chan et. al. 1 2020 . Though we are reasonably sure the genes here are well annotated, we want to note that not every protein has been verified to be expressed or functional during SARS-CoV-2 infections, either in vitro or in vivo. In an effort to be as comprehensive and transparent as possible, we are reporting the sub-networks of these functionally unverified proteins along with the other SARS-CoV-2 proteins. In such cases, we have made notes within the text below, and on the corresponding subnetwork figures, and would advise that more caution be taken when examining these proteins and their molecular interactions. Due to practical limits in our sample preparation and data collection process, we were unable to generate data for proteins corresponding to Nsp3, Orf7b, and Nsp16. Therefore these three genes have been left out of the following literature review of the SARS-CoV-2 proteins and the protein-protein interactions (PPIs) identified in this study. -

Symposium on Viral Membrane Proteins
Viral Membrane Proteins ‐ Shanghai 2011 交叉学科论坛 Symposium for Advanced Studies 第二十七期:病毒离子通道蛋白的结构与功能研讨会 Symposium on Viral Membrane Proteins 主办单位:中国科学院上海交叉学科研究中心 承办单位:上海巴斯德研究所 1 Viral Membrane Proteins ‐ Shanghai 2011 Symposium on Viral Membrane Proteins Shanghai Institute for Advanced Studies, CAS Institut Pasteur of Shanghai,CAS 30.11. – 2.12 2011 Shanghai, China 2 Viral Membrane Proteins ‐ Shanghai 2011 Schedule: Wednesday, 30th of November 2011 Morning Arrival Thursday, 1st of December 2011 8:00 Arrival 9:00 Welcome Bing Sun, Co-Director, Pasteur Institute Shanghai 9: 10 – 9:35 Bing Sun, Pasteur Institute Shanghai Ion channel study and drug target fuction research of coronavirus 3a like protein. 9:35 – 10:00 Tim Cross, Tallahassee, USA The proton conducting mechanism and structure of M2 proton channel in lipid bilayers. 10:00 – 10:25 Shy Arkin, Jerusalem, IL A backbone structure of SARS Coronavirus E protein based on Isotope edited FTIR, X-ray reflectivity and biochemical analysis. 10:20 – 10:45 Coffee Break 10:45 – 11:10 Rainer Fink, Heidelberg, DE Elektromechanical coupling in muscle: a viral target? 11:10 – 11:35 Yechiel Shai, Rehovot, IL The interplay between HIV1 fusion peptide, the transmembrane domain and the T-cell receptor in immunosuppression. 11:35 – 12:00 Christoph Cremer, Mainz and Heidelberg University, DE Super-resolution Fluorescence imaging of cellular and viral nanostructures. 12:00 – 13:30 Lunch Break 3 Viral Membrane Proteins ‐ Shanghai 2011 13:30 – 13:55 Jung-Hsin Lin, National Taiwan University Robust Scoring Functions for Protein-Ligand Interactions with Quantum Chemical Charge Models. 13:55 – 14:20 Martin Ulmschneider, Irvine, USA Towards in-silico assembly of viral channels: the trials and tribulations of Influenza M2 tetramerization. -

Opportunistic Intruders: How Viruses Orchestrate ER Functions to Infect Cells
REVIEWS Opportunistic intruders: how viruses orchestrate ER functions to infect cells Madhu Sudhan Ravindran*, Parikshit Bagchi*, Corey Nathaniel Cunningham and Billy Tsai Abstract | Viruses subvert the functions of their host cells to replicate and form new viral progeny. The endoplasmic reticulum (ER) has been identified as a central organelle that governs the intracellular interplay between viruses and hosts. In this Review, we analyse how viruses from vastly different families converge on this unique intracellular organelle during infection, co‑opting some of the endogenous functions of the ER to promote distinct steps of the viral life cycle from entry and replication to assembly and egress. The ER can act as the common denominator during infection for diverse virus families, thereby providing a shared principle that underlies the apparent complexity of relationships between viruses and host cells. As a plethora of information illuminating the molecular and cellular basis of virus–ER interactions has become available, these insights may lead to the development of crucial therapeutic agents. Morphogenesis Viruses have evolved sophisticated strategies to establish The ER is a membranous system consisting of the The process by which a virus infection. Some viruses bind to cellular receptors and outer nuclear envelope that is contiguous with an intri‑ particle changes its shape and initiate entry, whereas others hijack cellular factors that cate network of tubules and sheets1, which are shaped by structure. disassemble the virus particle to facilitate entry. After resident factors in the ER2–4. The morphology of the ER SEC61 translocation delivering the viral genetic material into the host cell and is highly dynamic and experiences constant structural channel the translation of the viral genes, the resulting proteins rearrangements, enabling the ER to carry out a myriad An endoplasmic reticulum either become part of a new virus particle (or particles) of functions5. -

Summary of Antimicrobial Activity
SUMMARY OF ANTIMICROBIAL ACTIVITY 3x RENEGADE DAILY ONE-STEP DISINFECTANT Description 3x RENEGADE DAILY Disinfectant & Detergent is a broad spectrum, hard surface disinfectant. When used as directed, this product will deliver effective biocidal action against bacteria, fungi, and viruses. This formulation is a blend of a premium active ingredients and inerts: surfactants, chelates, and water. Biocidal performance is attained when this product is properly diluted at 1/2 oz. per gallon or 1:256 (1 oz. per gallon or 1:128 for Norovirus). 3x RENEGADE DAILY can be used to disinfect a wide variety of hard surfaces such as floors, walls, toilets, sinks, and countertops in hospitals, households, and institutions. Regulatory Summary Physical Properties EPA Registration No. 6836-349- pH of Concentrate 12.0 – 13.5 Flash Point (PMCC) >200 F 12120 USDA Authorization None Specific Gravity @ 0.98 – 1.05 g/mL % Quat (mol. wt.342.0) 22.24 25°C California Status Pounds per gallon @ 8.42 – 8.51 % Volatile 93.5-94.5 25°C Canadian PCP# None Canadian Din # None Summary of Antimicrobial Test Results 3x RENEGADE DAILY is a "One-Step" Hospital Disinfectant, Virucide, Fungicide, Mildewstat, Sanitizer and Cleaner. Listed in the following pages is a summary of Antimicrobial Claims and a review of test results. Claim: Contact time: Organic Soil: Water Conditions: Disinfectant Varies 5% 250ppm as CaCO3 Test Method: AOAC Germicidal Spray Test Organism Contact Dilution Time (Min) 868 ppm (1/2oz. per Acinetobacter baumannii 3 Gal) Bordetella bronchiseptica 3 868 ppm Bordetella pertussis 3 868 ppm Campylobacter jejuni 3 868 ppm Enterobacter aerogenes 3 1736 PPM (1 oz per Gal) Enterococcus faecalis 3 868 ppm Enterococcus faecalis - Vancomycin resistant [VRE] 3 868 ppm Escherichia coli 3 868 ppm Escherichia coli [O157:H7] 3 868 ppm Escherichia coli ESBL – Extended spectrum beta- 868 ppm 10 lactamase containing E. -

Coronavirus Proteins As Ion Channels: Current and Potential Research
Washington University School of Medicine Digital Commons@Becker Open Access Publications 1-1-2020 Coronavirus proteins as ion channels: Current and potential research Conor McClenaghan Alex Hanson Sun-Joo Lee Colin G Nichols Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs REVIEW published: 09 October 2020 doi: 10.3389/fimmu.2020.573339 Coronavirus Proteins as Ion Channels: Current and Potential Research Conor McClenaghan, Alex Hanson, Sun-Joo Lee and Colin G. Nichols* Center for Investigation of Membrane Excitability Diseases, and Department of Cell Biology and Physiology, Washington University School of Medicine, St. Louis, MO, United States Coronavirus (CoV) outbreaks have recently emerged as a global public health threat due to their exceptional zoonotic potential — a feature arising from their ability to infect a diverse range of potential hosts combined with their high capacity for mutation and recombination. After Severe Acute Respiratory Syndrome (SARS) CoV-1 in 2003 and Middle East Respiratory Syndrome (MERS) CoV in 2012, with the current SARS-CoV-2 pandemic we are now in the midst of the third deadly international CoV outbreak in less than 20 years. Coronavirus outbreaks present a critical threat to global public health and Edited by: Julia Kzhyshkowska, an urgent necessity for therapeutic options. Here, we critically examine the current Heidelberg University, Germany evidence for ion channel activity in CoV proteins and the potential for modulation as a Reviewed by: therapeutic approach. Jaume Torres, Nanyang Technological University, Keywords: Severe Acute Respiratory Syndrome coronavirus-2, ion channel, spike protein, electrophysiology, Singapore bilayer, Severe Acute Respiratory Syndrome coronavirus Srinivasa Reddy Bonam, Institut National de la Sante´ et de la Recherche Me´ dicale (INSERM), France INTRODUCTION *Correspondence: Coronaviruses (CoVs) are enveloped, single-stranded, positive-sense RNA viruses that were first Colin G. -

Function and Structure of Viral Ribonucleoproteins Complexes”
viruses Editorial Special Issue “Function and Structure of Viral Ribonucleoproteins Complexes” Serena Bernacchi Architecture et Réactivité de l’ARN-CNRS UPR 9002, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, 67084 Strasbourg, France; [email protected] Academic Editor: Eric Freed Received: 23 November 2020; Accepted: 24 November 2020; Published: 26 November 2020 RNA viruses are extraordinary evolution machines that efficiently ensure their replication by taking advantage of the association with viral and cellular components to form ribonucleic complexes (vRNPs). These vRNPs are functional units of the infectious cycle, driving various processes including transcription, nuclear export, translation, and intracellular trafficking pathways for targeting viral components to the assembly sites where the packaging of the viral RNA genetic material into virions occurs. The aim of this Special Issue of Viruses is to provide an updated picture of the structural organization and of the different roles of vRNPs in various viral systems. The first part of the SI is dedicated to HIV-1 which is the causative agent of AIDS. Since its discovery in 1983, HIV-1 has become one of the leading causes of death worldwide. Up to now, the pandemic persists despite the implementation of highly active antiretroviral therapy, and over the years, a vast spectrum of techniques has been implemented to diagnose and monitor AIDS progression. Besides the conventional approaches, recently, microfluidics has provided useful methods for monitoring HIV-1 infection. This powerful tool allows information at the single-cell scale and high-throughput development. Herein, Eid et al. highlighted recent significant advances of continuous microfluidics in AIDS diagnosis, and in the basic study of the HIV-1 life cycle [1]. -

Risk Groups: Viruses (C) 1988, American Biological Safety Association
Rev.: 1.0 Risk Groups: Viruses (c) 1988, American Biological Safety Association BL RG RG RG RG RG LCDC-96 Belgium-97 ID Name Viral group Comments BMBL-93 CDC NIH rDNA-97 EU-96 Australia-95 HP AP (Canada) Annex VIII Flaviviridae/ Flavivirus (Grp 2 Absettarov, TBE 4 4 4 implied 3 3 4 + B Arbovirus) Acute haemorrhagic taxonomy 2, Enterovirus 3 conjunctivitis virus Picornaviridae 2 + different 70 (AHC) Adenovirus 4 Adenoviridae 2 2 (incl animal) 2 2 + (human,all types) 5 Aino X-Arboviruses 6 Akabane X-Arboviruses 7 Alastrim Poxviridae Restricted 4 4, Foot-and- 8 Aphthovirus Picornaviridae 2 mouth disease + viruses 9 Araguari X-Arboviruses (feces of children 10 Astroviridae Astroviridae 2 2 + + and lambs) Avian leukosis virus 11 Viral vector/Animal retrovirus 1 3 (wild strain) + (ALV) 3, (Rous 12 Avian sarcoma virus Viral vector/Animal retrovirus 1 sarcoma virus, + RSV wild strain) 13 Baculovirus Viral vector/Animal virus 1 + Togaviridae/ Alphavirus (Grp 14 Barmah Forest 2 A Arbovirus) 15 Batama X-Arboviruses 16 Batken X-Arboviruses Togaviridae/ Alphavirus (Grp 17 Bebaru virus 2 2 2 2 + A Arbovirus) 18 Bhanja X-Arboviruses 19 Bimbo X-Arboviruses Blood-borne hepatitis 20 viruses not yet Unclassified viruses 2 implied 2 implied 3 (**)D 3 + identified 21 Bluetongue X-Arboviruses 22 Bobaya X-Arboviruses 23 Bobia X-Arboviruses Bovine 24 immunodeficiency Viral vector/Animal retrovirus 3 (wild strain) + virus (BIV) 3, Bovine Bovine leukemia 25 Viral vector/Animal retrovirus 1 lymphosarcoma + virus (BLV) virus wild strain Bovine papilloma Papovavirus/ -

Immunodeficiency Virus Genome Affects Virus Production and Infectivity T
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 4114-4118, May 1992 Microbiology Mutation in the primer binding site of the type 1 human immunodeficiency virus genome affects virus production and infectivity T. NAGASHUNMUGAM*, A. VELPANDI*, C. S. GOLDSMITHt, S. R. ZAKIt, V. S. KALYANARAMANt, AND A. SRINIVASAN*§ *The Wistar Institute of Anatomy and Biology, 3601 Spruce Street, Philadelphia, PA 19104; tDivision of Viral and Rickettsial Diseases, Center for Infectious Diseases, Centers for Disease Control, Atlanta, GA 30333; and tAdvanced Bioscience Laboratories, Inc., 5510 Nicholson Lane, Kensington, MD 20805 Communicated by Hilary Koprowski, January 16, 1992 ABSTRACT In an effort to understand the contribution of retroviruses make use of tryptophan tRNA. Sequence anal- the primer-binding site (PBS) region to human immunodefi- ysis of HIV-1 revealed PBS corresponding to the lysine ciency virus (HIV) replication, we have constructed a mutant tRNA as in other lentiviruses (7). It has also been recently HIV proviral DNA with an alteration in the 5' end of the PBS. demonstrated that HIV RT forms a stable complex with The PBS mutant proviral DNA was characterized by transfec- isoacceptor 3 of lysine tRNA (Lys3 tRNA) (8), as has been tion of the viral DNA into CD4' and non-CD4' target cells. demonstrated for the RT enzyme of avian viruses. The results indicate that mutation in the PBS reduced the level Initiation of viral DNA synthesis is a crucial step in the of viral particles released into the medium of transfected cells replication of retroviruses, and the essential PBS is embed- in comparison to wild-type proviral DNA. -

Arenaviridae Astroviridae Filoviridae Flaviviridae Hantaviridae
Hantaviridae 0.7 Filoviridae 0.6 Picornaviridae 0.3 Wenling red spikefish hantavirus Rhinovirus C Ahab virus * Possum enterovirus * Aronnax virus * * Wenling minipizza batfish hantavirus Wenling filefish filovirus Norway rat hunnivirus * Wenling yellow goosefish hantavirus Starbuck virus * * Porcine teschovirus European mole nova virus Human Marburg marburgvirus Mosavirus Asturias virus * * * Tortoise picornavirus Egyptian fruit bat Marburg marburgvirus Banded bullfrog picornavirus * Spanish mole uluguru virus Human Sudan ebolavirus * Black spectacled toad picornavirus * Kilimanjaro virus * * * Crab-eating macaque reston ebolavirus Equine rhinitis A virus Imjin virus * Foot and mouth disease virus Dode virus * Angolan free-tailed bat bombali ebolavirus * * Human cosavirus E Seoul orthohantavirus Little free-tailed bat bombali ebolavirus * African bat icavirus A Tigray hantavirus Human Zaire ebolavirus * Saffold virus * Human choclo virus *Little collared fruit bat ebolavirus Peleg virus * Eastern red scorpionfish picornavirus * Reed vole hantavirus Human bundibugyo ebolavirus * * Isla vista hantavirus * Seal picornavirus Human Tai forest ebolavirus Chicken orivirus Paramyxoviridae 0.4 * Duck picornavirus Hepadnaviridae 0.4 Bildad virus Ned virus Tiger rockfish hepatitis B virus Western African lungfish picornavirus * Pacific spadenose shark paramyxovirus * European eel hepatitis B virus Bluegill picornavirus Nemo virus * Carp picornavirus * African cichlid hepatitis B virus Triplecross lizardfish paramyxovirus * * Fathead minnow picornavirus -
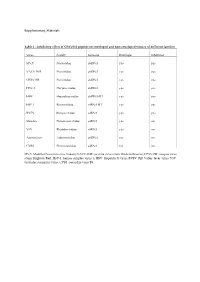
Inhibitory Effect of CPXV012 Peptide on Enveloped and Non-Enveloped Viruses of Different Families
Supplementary Materials Table 1 - Inhibitory effect of CPXV012 peptide on enveloped and non-enveloped viruses of different families Virus Family Genome Envelope Inhibition MVA Poxviridae dsDNA yes yes VACV-WR Poxviridae dsDNA yes yes CPXV-BR Poxviridae dsDNA yes yes HSV-1 Herpesviridae dsDNA yes yes HBV Hepadnaviridae dsDNA-RT yes yes HIV-1 Retroviridae ssRNA-RT yes yes RVFV Bunyaviridae ssRNA yes yes Measles Paramyxoviridae ssRNA yes no VSV Rhabdoviridae ssRNA yes no Adenovirus Adenoviridae dsDNA no no CVB3 Picornaviridae ssRNA no no MVA: Modified Vaccinia virus Ankara; VACV-WR: vaccinia virus strain Western Reserve; CPXV-BR: cowpox virus strain Brighton Red; HSV-1: herpes simplex virus-1; HBV: Hepatitis B virus; RVFV: Rift Valley fever virus; VSV: vesicular stomatitis virus; CVB3: coxsackie virus B3. A B 140 0.6 ) l 120 ro t 100 on 0.4 c g f 80 µ / o 60 % NR ( 0.2 1 - 40 T S 20 W 0 0.0 O 5 0 0 0 5 0 0 0 S 2 5 0 0 O 2 5 0 0 1 2 S 1 2 M M D D CPXV012 peptide [ g/mL] CPXV012 peptide [ g/mL] C ) . 30000 U . A ( e u 20000 l b r e t i t 10000 ll e C 0 0 50 100 150 200 CPXV012 peptide [ g/mL] Huh7.5 Vero Figure S1: CPXV012 peptide does not affect cell viability. (A-C) Cells were treated with the indicated concentrations of peptide or DMSO as vehicle control. S.E.M. of three independent experiments is shown. (A) Viability of MJS cells was measured by WST-1 assay or (B) Neutral Red uptake.