Polynesian Plant Introductions in the Southwest Pacific: Initial Pollen Evidence from Norfolk Island
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
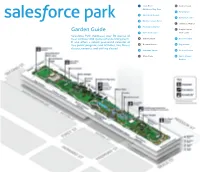
Salesforce Park Garden Guide
Start Here! D Central Lawn Children’s Play Area Garden Guide6 Palm Garden 1 Australian Garden Start Here! D Central Lawn Salesforce Park showcases7 California over Garden 50 species of Children’s Play Area 2 Mediterraneantrees and Basin over 230 species of understory plants. 6 Palm Garden -ã ¼ÜÊ ÊăØÜ ØÊèÜãE úØƀØÊèÃJapanese Maples ¼ÃØ Ê¢ 1 Australian Garden 3 Prehistoric¢ØÕ輫ÕØÊ£ØÂÜÃã«ó«ã«Üŧ¼«¹ĆãÃÜÜ Garden 7 California Garden ¼ÜÜÜŧÊÃØãÜŧÃØ¢ã«Ã£¼ÜÜÜũF Amphitheater Garden Guide 2 Mediterranean Basin 4 Wetland Garden Main Lawn E Japanese Maples Salesforce Park showcases over 50 species of 3 Prehistoric Garden trees and over 230 species of understory plants. A Oak Meadow 8 Desert Garden F Amphitheater It also offers a robust year-round calendar of 4 Wetland Garden Main Lawn free public programs and activities, like fitness B Bamboo Grove 9 Fog Garden Desert Garden classes, concerts, and crafting classes! A Oak Meadow 8 5 Redwood Forest 10 Chilean Garden B Bamboo Grove 9 Fog Garden C Main Plaza 11 South African 10 Chilean Garden Garden 5 Redwood Forest C Main Plaza 11 South African Garden 1 Children’s Australian Play Area Garden ABOUT THE GARDENS The botanist aboard the Endeavor, Sir Joseph Banks, is credited with introducing many plants from Australia to the western world, and many This 5.4 acre park has a layered soil system that plants today bear his name. balances seismic shifting, collects and filters storm- water, and irrigates the gardens. Additionally, the soil Native to eastern Australia, Grass Trees may grow build-up and dense planting help offset the urban only 3 feet in 100 years, and mature plants can be heat island effect by lowering the air temperature. -

Approaching the Prehistory of Norfolk Island
© Copyright Australian Museum, 2001 Records of the Australian Museum, Supplement 27 (2001): 1–9. ISBN 0 7347 2305 9 Approaching the Prehistory of Norfolk Island ATHOLL ANDERSON1 AND PETER WHITE2 1 Department of Archaeology & Natural History, Research School of Pacific and Asian Studies, Australian National University, Canberra ACT 0200, Australia [email protected] 2 Archaeology, University of Sydney, Sydney NSW 2006, Australia [email protected] ABSTRACT. Norfolk Island, on the northeast edge of the Tasman Sea, is of volcanic origin and moderate height. A humid, forested subtropical landmass, it had a diverse range of natural resources, including some food plants such as Cyathea, forest birds such as pigeon and parrot species and substantial colonies of seabirds, notably boobies and procellariids. Its shoreline had few shellfish, but the coastal waters were rich in fish, of which Lethrinids were especially abundant. The island had no inhabitants when discovered by Europeans in A.D. 1774. It was settled by them in A.D. 1788. From the eighteenth century discovery of feral bananas and then of stone adzes, knowledge of the prehistory of Norfolk Island has developed over a very long period. Collections of stone tools seemed predominantly East Polynesian in orientation, but Melanesian sources could not be ruled out. Research on fossil bone deposits established the antiquity of the human commensal Rattus exulans as about 800 B.P. but no prehistoric settlement site was known until one was discovered in 1995 at Emily Bay during the Norfolk Island Prehistory Project. ANDERSON, ATHOLL, AND PETER WHITE, 2001a. Approaching the prehistory of Norfolk Island. -

TAXON:Rhopalostylis Baueri SCORE:-2.0 RATING:Low Risk
TAXON: Rhopalostylis baueri SCORE: -2.0 RATING: Low Risk Taxon: Rhopalostylis baueri Family: Arecaceae Common Name(s): Norfolk Island palm Synonym(s): Areca baueri Hook. f. ex Lem. Eora(basionym) baueri (H. Wendl. & Drude) O. F. RhopalostylisCook cheesemanii Becc. ex Cheeseman Assessor: No Assessor Status: Assessor Approved End Date: WRA Score: -2.0 Designation: L Rating: Low Risk Keywords: Subtropical Palm, Unarmed, Shade-tolerant, Thicket-forming, Bird-dispersed Qsn # Question Answer Option Answer 101 Is the species highly domesticated? y=-3, n=0 n 102 Has the species become naturalized where grown? 103 Does the species have weedy races? Species suited to tropical or subtropical climate(s) - If 201 island is primarily wet habitat, then substitute "wet (0-low; 1-intermediate; 2-high) (See Appendix 2) High tropical" for "tropical or subtropical" 202 Quality of climate match data (0-low; 1-intermediate; 2-high) (See Appendix 2) High 203 Broad climate suitability (environmental versatility) y=1, n=0 n Native or naturalized in regions with tropical or 204 y=1, n=0 y subtropical climates Does the species have a history of repeated introductions 205 y=-2, ?=-1, n=0 y outside its natural range? 301 Naturalized beyond native range y = 1*multiplier (see Appendix 2), n= question 205 n 302 Garden/amenity/disturbance weed n=0, y = 1*multiplier (see Appendix 2) n 303 Agricultural/forestry/horticultural weed n=0, y = 2*multiplier (see Appendix 2) n 304 Environmental weed n=0, y = 2*multiplier (see Appendix 2) n 305 Congeneric weed n=0, y = 1*multiplier -

Rhopalostylis Sapida
Rhopalostylis sapida COMMON NAME Nikau palm SYNONYMS None FAMILY Arecaceae AUTHORITY Rhopalostylis sapida H.Wendl. et Drude FLORA CATEGORY Vascular – Native ENDEMIC TAXON Yes ENDEMIC GENUS No ENDEMIC FAMILY No STRUCTURAL CLASS Trees & Shrubs - Monocotyledons NVS CODE RHOSAP CHROMOSOME NUMBER Whareroa Farm, Paekakariki. Apr 2011. 2n = 32 Photographer: Jeremy Rolfe CURRENT CONSERVATION STATUS 2012 | Not Threatened PREVIOUS CONSERVATION STATUSES 2009 | Not Threatened 2004 | Not Threatened BRIEF DESCRIPTION Palm to 15m tall with a ringed trunk and 3m long erect leaves inhabiting lowland forest south to Okarito and Banks Peninsula and the Chatham Islands. Leaves with multiple narrow leaflets to 1m long closely-spaced along central stem. Flowers pinkish, in multiple spikes at the top of trunk. Fruit red. DISTRIBUTION Endemic. North Island, South Island from Marlborough Sounds and Nelson south to Okarito in the west and Banks Peninsula in the east. Also on Chatham and Pitt Islands. However Chatham Islands plants have adistinct juveniel form, larger fruits, and thicker indumentum on the fronds. HABITAT Trunk of nikau. Photographer: Wayne Bennett Primarily a species of coastal to lowland forest in the warmer parts of New Zealand. FEATURES Trunk up to 15 m, stout, covered in grey-green leaf scars, otherwise green. Crownshaft 0.6(-1) m long, dark green, smooth, bulging. Fronds up to 3 m long; leaflets to 1 m, closely set (sometimes over lapping), ascending. Spathes c.300 x 150 mm., between pink and yellow, caducous. Inflorescence shortly stalked, with many branches, 200-400 mm long. Flowers sessile, unisexual, tightly packed, lilac to pink. Males in pairs, caducous, stamens 6. -

The Island Rule and Its Application to Multiple Plant Traits
The island rule and its application to multiple plant traits Annemieke Lona Hedi Hendriks A thesis submitted to the Victoria University of Wellington in partial fulfilment of the requirements for the degree of Master of Science in Ecology and Biodiversity Victoria University of Wellington, New Zealand 2019 ii “The larger the island of knowledge, the longer the shoreline of wonder” Ralph W. Sockman. iii iv General Abstract Aim The Island Rule refers to a continuum of body size changes where large mainland species evolve to become smaller and small species evolve to become larger on islands. Previous work focuses almost solely on animals, with virtually no previous tests of its predictions on plants. I tested for (1) reduced floral size diversity on islands, a logical corollary of the island rule and (2) evidence of the Island Rule in plant stature, leaf size and petiole length. Location Small islands surrounding New Zealand; Antipodes, Auckland, Bounty, Campbell, Chatham, Kermadec, Lord Howe, Macquarie, Norfolk, Snares, Stewart and the Three Kings. Methods I compared the morphology of 65 island endemics and their closest ‘mainland’ relative. Species pairs were identified. Differences between archipelagos located at various latitudes were also assessed. Results Floral sizes were reduced on islands relative to the ‘mainland’, consistent with predictions of the Island Rule. Plant stature, leaf size and petiole length conformed to the Island Rule, with smaller plants increasing in size, and larger plants decreasing in size. Main conclusions Results indicate that the conceptual umbrella of the Island Rule can be expanded to plants, accelerating understanding of how plant traits evolve on isolated islands. -

Studies on Industrially Important Guttiferae and Palmae Family
Journal of Pharmacognosy and Phytochemistry 2016; 5(6): 194-198 E-ISSN: 2278-4136 P-ISSN: 2349-8234 Studies on industrially important Guttiferae and JPP 2016; 5(6): 194-198 Palmae family Received: 18-05-2016 Accepted: 19-06-2016 Kembavimath M Kotraswamy Kembavimath M Kotraswamy, Irfan N Shaikh, Rajasaheb F Ankalgi, Department of Chemistry, G.S. Shamsunnisa R Ankalgi, Imran N Shaikh and Umar Farooq Bagwan Science College, Belgaum, India Abstract Irfan N Shaikh Department of Chemistry, The Garcinia mangostana seed oil contains 56.2% oleic acid and 6.4% linoleic acid. The palmitic SECAB Institute of Engineering (14.8%) and stearic acid (9.0%) are major components amongst the saturated acids with smaller amounts & Technology, Vijayapura, India of capric (0.9%), lauric (2.2%), myristic (6.6%) and arachidic (3.9%). Moreover, Phoenix sylvestris, cerita mistis, chrysalidocarpus lutescens, Washingtonia filifera and phoenix rupicola belong to palmae Rajasaheb F Ankalgi family and could be compared with the oils rich in lauric acid such as cinnamon and palm kernel oils (80- Essar Laboratories and Research 90% and 45-58% respectively). Centre, Hubli, India Keywords: Guttiferae, Palmae, fatty acid, industrially important Shamsunnisa R Ankalgi Essar Laboratories and Research 1. Introduction Centre, Hubli, India One of the important facts of plants is their diverse pool of fatty acids. The oil seeds contains Imran N Shaikh particular fatty acids with industrially important because of their characteristic properties. The Department of Chemistry, main constituent of all the oils is the fatty acids which may include saturated, monounsaturated Mahatma Gandhi PU Science and polyunsaturated fatty acid that contribute in human physiology in different ways [1]. -

(Arecaceae): Évolution Du Système Sexuel Et Du Nombre D'étamines
Etude de l’appareil reproducteur des palmiers (Arecaceae) : évolution du système sexuel et du nombre d’étamines Elodie Alapetite To cite this version: Elodie Alapetite. Etude de l’appareil reproducteur des palmiers (Arecaceae) : évolution du système sexuel et du nombre d’étamines. Sciences agricoles. Université Paris Sud - Paris XI, 2013. Français. NNT : 2013PA112063. tel-01017166 HAL Id: tel-01017166 https://tel.archives-ouvertes.fr/tel-01017166 Submitted on 2 Jul 2014 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITE PARIS-SUD ÉCOLE DOCTORALE : Sciences du Végétal (ED 45) Laboratoire d'Ecologie, Systématique et E,olution (ESE) DISCIPLINE : -iologie THÈSE DE DOCTORAT SUR TRAVAUX soutenue le ./05/10 2 par Elodie ALAPETITE ETUDE DE L'APPAREIL REPRODUCTEUR DES PAL4IERS (ARECACEAE) : EVOLUTION DU S5STE4E SE6UEL ET DU NO4-RE D'ETA4INES Directeur de thèse : Sophie NADOT Professeur (Uni,ersité Paris-Sud Orsay) Com osition du jury : Rapporteurs : 9ean-5,es DU-UISSON Professeur (Uni,ersité Pierre et 4arie Curie : Paris VI) Porter P. LOWR5 Professeur (4issouri -otanical Garden USA et 4uséum National d'Histoire Naturelle Paris) Examinateurs : Anders S. -ARFOD Professeur (Aarhus Uni,ersity Danemark) Isabelle DA9OA Professeur (Uni,ersité Paris Diderot : Paris VII) 4ichel DRON Professeur (Uni,ersité Paris-Sud Orsay) 3 4 Résumé Les palmiers constituent une famille emblématique de monocotylédones, comprenant 183 genres et environ 2500 espèces distribuées sur tous les continents dans les zones tropicales et subtropicales. -

WUCOLS List S Abelia Chinensis Chinese Abelia M ? ? M / / Copyright © UC Regents, Davis Campus
Ba Bu G Gc P Pm S Su T V N Botanical Name Common Name 1 2 3 4 5 6 Symbol Vegetation Used in Type WUCOLS List S Abelia chinensis Chinese abelia M ? ? M / / Copyright © UC Regents, Davis campus. All rights reserved. bamboo Ba S Abelia floribunda Mexican abelia M ? M M / / S Abelia mosanensis 'Fragrant Abelia' fragrant abelia ? ? ? ? ? ? bulb Bu S Abelia parvifolia (A. longituba) Schuman abelia ? ? ? M ? ? grass G groundcover GC Gc S Abelia x grandiflora and cvs. glossy abelia M M M M M / perennial* P S Abeliophyllum distichum forsythia M M ? ? ? ? palm and cycad Pm S Abelmoschus manihot (Hibiscus manihot) sunset muskmallow ? ? ? L ? ? T Abies pinsapo Spanish fir L L L / / / shrub S succulent Su T N Abies spp. (CA native and non-native) fir M M M M / / P N Abronia latifolia yellow sand verbena VL VL VL / ? ? tree T P N Abronia maritima sand verbena VL VL VL / ? ? vine V California N native S N Abutilon palmeri Indian mallow L L L L M M S Abutilon pictum thompsonii variegated Chinese lantern M H M M ? ? Sunset WUCOLS CIMIS ET Representative Number climate 0 Region zones** Cities zones* S Abutilon vitifolium flowering maple M M M / ? ? Healdsburg, Napa, North- San Jose, Salinas, Central 14, 15, 16, 17 1, 2, 3, 4, 6, 8 San Francisco, Coastal San Luis Obispo S Abutilon x hybridum & cvs. flowering maple M H M M / / 1 Auburn, Central Bakersfield, Chico, 8, 9, 14 12, 14, 15, 16 Valley Fresno, Modesto, Sacramento S T Acacia abyssinica Abyssinian acacia / ? / ? / L 2 Irvine, Los South Angeles, Santa 22, 23, 24 1, 2, 4, 6 Coastal Barbara, Ventura, -

The Kermadec Islands Terrestrial Invertebrate Fauna: Observations on the Taxonomic Distribution and Island Biogeography
www.aucklandmuseum.com The Kermadec Islands terrestrial invertebrate fauna: Observations on the taxonomic distribution and island biogeography Warren G.H. Chinn Department of Conservation Abstract A sample of terrestrial invertebrates from the Kermadec Islands is reported on. Specimens were acquired as part of a marine biodiversity expedition in May 2011, consisting of: 118 recognisable taxonomic units; 12 unrecognised taxa and 47 species with new location records. Seven of the 15 Kermadec Islands were visited, including Raoul, the twin Meyers, North Chanter, Macauley, Cheeseman and L’Esperance Rock. Of these, only Raoul, the Meyers and Macauley Islands have received previous entomological attention. The level of endemism is extremely low, and most of the Kermadec Island fauna is also found elsewhere in the south Pacific which comprises highly mobile taxa. The taxonomic composition of the four most isolated islands is examined by class and shows a reduction of groups associated with land area. A species area curve suggests the Kermadec Islands are consistent with the theory of island biogeography, demonstrating a positive relationship (r2=0.4538) between the number of taxa and island size. Inter-island dispersal is examined using the percentage of shared taxa correlated with distance between islands. A weak negative relationship (-0.164) suggests that distance is a minor barrier, while habitat suitability may be more significant and could operate as an ecological filter. These interpretations highlight the value of the island group as a biogeography laboratory while reinforcing their conservation value. Keywords Kermadec Islands; Raoul Island; Macauley Island; Cheeseman Island; Curtis Island; L’Esperance Rock; invertebrate fauna; diptera; araneae; orthoptera; island biogeography; species area relationship INTRODUCTION ngaio (Myoporum rapense subsp. -

Norfolk Island Region Threatened Species Recovery Plan
NORFOLK ISLAND Norfolk Island Region CouncilThreatened of Heads of Australian Species Botanic GardensRecovery Plan November 2008 Prepared by: Director of National Parks Made under the Environment Protection and Biodiversity Conservation Act 1999 ISBN: 978-0-646-53763-4 © Commonwealth of Australia 2010 This work is copyright. You may download, display, print and reproduce this material in unaltered form only (retaining this notice) for your personal, non-commercial use or use within your organisation. Apart from any use as permitted under the Copyright Act 1968, all other rights are reserved. Requests and inquiries concerning reproduction and rights should be addressed to Commonwealth Copyright Administration, Attorney General’s Department, Robert Garran Offices, National Circuit, Barton ACT 2600 or posted at: ag.gov.au/cca Note: This recovery plan sets out the actions necessary to stop the decline of, and support the recovery of, listed threatened species. The Australian Government is committed to acting in accordance with the plan and to implementing the plan as it applies to Commonwealth areas. The plan has been developed with the involvement and cooperation of a broad range of stakeholders, but individual stakeholders have not necessarily committed to undertaking specific actions. The attainment of objectives and the provision of funds is subject to budgetary and other constraints affecting the parties involved. Proposed actions may be subject to modification over the life of the plan due to changes in knowledge. While reasonable efforts have been made to ensure that the contents of this publication are factually correct, the Commonwealth does not accept responsibility for the accuracy or completeness of the contents, and shall not be liable for any loss or damage that may be occasioned directly or indirectly through the use of, or reliance on, the contents of this publication. -

Ancient Polyploidy and Genome Evolution in Palms
Faculty & Staff Scholarship 2019 Ancient Polyploidy and Genome Evolution in Palms Craig F. Barrett Michael R. McKain Brandon T. Sinn Xue Jun Ge Yuqu Zhang See next page for additional authors Follow this and additional works at: https://researchrepository.wvu.edu/faculty_publications Part of the Biology Commons Authors Craig F. Barrett, Michael R. McKain, Brandon T. Sinn, Xue Jun Ge, Yuqu Zhang, Alexandre Antonelli, and Christine D. Bacon GBE Ancient Polyploidy and Genome Evolution in Palms Craig F. Barrett1,*, Michael R. McKain2, Brandon T. Sinn1, Xue-Jun Ge3, Yuqu Zhang3, Alexandre Antonelli4,5,6, and Christine D. Bacon4,5 1Department of Biology, West Virginia University 2Department of Biological Sciences, University of Alabama 3Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, PR China Downloaded from https://academic.oup.com/gbe/article-abstract/11/5/1501/5481000 by guest on 05 May 2020 4Department of Biological and Environmental Sciences, University of Gothenburg, Sweden 5Gothenburg Global Biodiversity Centre, Go¨ teborg, Sweden 6Royal Botanical Gardens Kew, Richmond, United Kingdom *Corresponding author: E-mail: [email protected]. Accepted: April 19, 2019 Data deposition: This project has been deposited at the NCBI Sequence Read Archive under the accession BioProject (PRJNA313089). Abstract Mechanisms of genome evolution are fundamental to our understanding of adaptation and the generation and maintenance of biodiversity, yet genome dynamics are still poorly characterized in many clades. Strong correlations between variation in genomic attributes and species diversity across the plant tree of life suggest that polyploidy or other mechanisms of genome size change confer selective advantages due to the introduction of genomic novelty. -

Guide to Estimating Irrigation Water Needs of Landscape Plantings in California
AA GuideGuide toto EstimatingEstimating IrrigationIrrigation WaterWater NeedsNeeds ofof LandscapeLandscape PlantingsPlantings inin CaliforniaCalifornia TheThe LandscapeLandscape CoefficientCoefficient MethodMethod andand WUCOLSWUCOLS IIIIII UniversityUniversity ofof CaliforniaCalifornia CooperativeCooperative ExtensionExtension CaliforniaCalifornia DepartmentDepartment ofof WaterWater ResourcesResources Cover photo: The Garden at Heather Farms, Walnut Creek, CA This Guide is a free publication. Additional copies may be obtained from: Department of Water Resources Bulletins and Reports P. O. Box 942836 Sacramento, California 94236-0001 (916) 653-1097 Photography: L.R. Costello and K.S. Jones, University of California Cooperative Extension Publication Design: A.S. Dyer, California Department of Water Resources A Guide to Estimating Irrigation Water Needs of Landscape Plantings in California The Landscape Coefficient Method and WUCOLS III* *WUCOLS is the acronym for Water Use Classifications of Landscape Species. University of California Cooperative Extension California Department of Water Resources August 2000 Preface This Guide consists of two parts, each formerly a ter needs for individual species. Used together, they separate publication: provide the information needed to estimate irriga- tion water needs of landscape plantings. Part 1—Estimating the Irrigation Water Needs of Landscape Plantings in California: The Land- Part 1 is a revision of Estimating Water Require- scape Coefficient Method ments of Landscape Plants: The Landscape Co- • L.R. Costello, University of California Coopera- efficient Method, 1991 (University of California tive Extension ANR Leaflet No. 21493). Information presented in • N.P. Matheny, HortScience, Inc., Pleasanton, CA the original publication has been updated and ex- • J.R. Clark, HortScience Inc., Pleasanton, CA panded. Part 2—WUCOLS III (Water Use Classification Part 2 represents the work of many individuals and of Landscape Species) was initiated and supported by the California De- • L.R.