Educational Workshop
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biofire Blood Culture Identification System (BCID) Fact Sheet
BioFire Blood Culture Identification System (BCID) Fact Sheet What is BioFire BioFire BCID is a multiplex polymerase chain reaction (PCR) test designed to BCID? identify 24 different microorganism targets and three antibiotic resistance genes from positive blood culture bottles. What is the purpose The purpose of BCID is to rapidly identify common microorganisms and of BCID? antibiotic resistance genes from positive blood cultures so that antimicrobial therapy can be quickly optimized by the physician and the antibiotic stewardship pharmacist. It is anticipated that this will result in improved patient outcomes, decreased length of stay, improved antibiotic stewardship, and decreased costs. When will BCID be BCID is performed on all initially positive blood cultures after the gram stain is routinely performed and reported. performed? When will BCID not For blood cultures on the same patient that subsequently become positive with be routinely a microorganism showing the same morphology as the initial positive blood performed? culture, BCID will not be performed. BCID will not be performed on positive blood cultures with gram positive bacilli unless Listeria is suspected. BCID will not be performed on blood culture bottles > 8 hours after becoming positive. BCID will not be performed between 10PM-7AM on weekdays and 2PM-7AM on weekends. BCID will not be performed for clinics that have specifically opted out of testing. How soon will BCID After the blood culture becomes positive and the gram stain is performed and results be available? reported, the bottle will be sent to the core Microbiology lab by routine courier. BCID testing will then be performed. It is anticipated that total turnaround time will generally be 2-3 hours after the gram stain is reported. -

Table 3. Distribution of Tetracycline Resistance Genes Among Gram-Positive Bacteria, Mycobacterium, Mycoplasma, Nocardia, Streptomyces and Ureaplasma Modified Sept
Table 3. Distribution of tetracycline resistance genes among Gram-positive bacteria, Mycobacterium, Mycoplasma, Nocardia, Streptomyces and Ureaplasma Modified Sept. 27, 2021 [n=58 genera] Originally modified from MMBR 2001; 65:232-260 with permission from ASM Journals One Determinant Two Determinants Three or More Determinants n=27 n=7 n=22 k Abiotrophia tet(M) Arthrobacter tet(33)(M) Actinomyces tet(L)(M)(W) Afipia tet(M) Gardnerella tet(M)(Q) Aerococcus tet(M)(O)(58)(61) o Amycolatopsis tet(M) Gemella tet(M)(O) Bacillus tet(K)(L)(M)(O)ao(T)ao(W)(39)m(42)I (45)atotr(A)L Anaerococcus tet(M)g Granulicatella tet(M)(O) Bifidobacterium a, w tet(L)(M)(O)(W) Bacterionema tet(M) Lactococcus tet(M)(S) Bhargavaea tet(L)ac(M)(45)aa ar Brachybacterium tet(M)k Mobiluncusa tet(O)(Q) Clostridiuma,f tet(K)(L)(M)(O)(P)(Q)(W)(36)(40)j(44)p(X) Catenibacteriuma tet(M) Savagea tet(L)(M) Clostridioidesat tet(L)(P)(W)(40) Cellulosimicrobium tet(39)m Corynebacterium tet(M)(Z)(33)(W)q (39)ak Cottaibacterium tet(M) Enterococcus tet(K)(L)(M)(O)(S)(T)(U)(58)ad(61)aq Cutibacterium tet(W)aq Eubacteriuma tet(K)(M)(O)(Q)(32) Erysipelothrix tet(M) Lactobacillusf tet(K)(L)(M)(O)(Q)(S)(W)(Z)(36)am Finegoldia tet(M)g Listeria tet(K)(L)(M)(S)AB(46)ag Geobacillus tet(L) Microbacterium tet(M)(O)ae(42)I Helcococcus tet(M)ah Mycobacteriumc tet(K)(L)(M)(O)t(V)arotr(A)(B) Leifsonia tet(O)t Nocardia tet(K)(L)(M)ai (O) ai Lysinibacillus tet(39)m Paenibacillus tet(L)(M)(O)t(42)i Micrococcus tet(42) Peptostreptococcusa tet(K)(L)(M)(O)(Q) Mycoplasmab tet(M) Sporosarcina tet(K)(L)ac(M)n -

Potential Role of Microbiome in Oncogenesis, Outcome Prediction
Oral Oncology 99 (2019) 104453 Contents lists available at ScienceDirect Oral Oncology journal homepage: www.elsevier.com/locate/oraloncology Review Potential role of microbiome in oncogenesis, outcome prediction and therapeutic targeting for head and neck cancer T ⁎ Ester Orlandia,b, , Nicola Alessandro Iacovellib, Vincenzo Tombolinic, Tiziana Rancatid, Antonella Polimenie, Loris De Ceccof, Riccardo Valdagnia,d,g, Francesca De Felicec a Department of Radiotherapy 1, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy b Department of Radiotherapy 2, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy c Department of Radiotherapy, Policlinico Umberto I, “Sapienza” University of Rome, Rome, Italy d Prostate Cancer Program, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy e Department of Oral and Maxillo Facial Sciences, Policlinico Umberto I, “Sapienza” University of Rome, Italy f Integrated Biology Platform, Department of Applied Research and Technology Development, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy g Department of Oncology and Hemato-Oncology, University of Milan, Milan, Italy ARTICLE INFO ABSTRACT Keywords: In the last decade, human microbiome research is rapidly growing involving several fields of clinical medicine Head and neck cancer and population health. Although the microbiome seems to be linked to all sorts of diseases, cancer has the Biomarkers biggest potential to be investigated. Microbiome Following the publication of the National Institute of Health - Human Microbiome Project (NIH-HMP), the Microbiota link between Head and Neck Cancer (HNC) and microbiome seems to be a fast-moving field in research area. Oncogenesis However, robust evidence-based literature is still quite scarce. Nevertheless the relationship between oral mi- Radiotherapy Immunotherapy crobiome and HNC could have important consequences for prevention and early detection of this type of tumors. -

3. Jedna Z Krvných Skupín Systému AB0; 4. Skr. Bukálny. B – Symbo
B – 1. symbol pre bel; 2. chem. značka prvku →bór; 3. jedna z krvných skupín systému AB0; 4. skr. bukálny. B – symbol pre hustotu magnetického toku. B. – skr. pre →Bacillus. B-ALP – skr. angl. bone alcalic phosphatase kostná alkalická fosfatáza. B-bunky – 1. syn. -bunky Langerhansových ostrovčekov; →pankreas; 2. syn. B-lymfocyty; →lymfocyty. B-komplex – multivitamínový prípravok vitamínov B. B-lymfocyty – [B podľa Fabriciovej burzy, imunol. orgánu vtákov] B bunky, druh lymfocytov, kt. sa zúčastňuje na humorálnej imunite (tvorbe protilátok) a niekt. ďalších imunitných funkciách. Povrchové molekuly sú CD 19,20. Receptorom pre antigén je membránový imunoglobulín. B-reťazec – jeden z polypeptidových reťazcov →inzulínu. B-vírus – 1. vírus →hepatitídy B.; 2. vírus zo skupiny Cercopithecine herpes virus 1. B-vlákna →nerv. b – skr. 1. pre barn; 2. skr. angl. born narodený; 3. skr. báza (genet. označenie dĺţky sekvencie nukleotidov, napr. 50 b = sekvencia 50 nukleotidov). b-vlna – pozit. kmit s vyskou amplitúdou v →elektroretinograme nasledujúci po vlne a, prejav komplexnej aktivity vrstvy bipolárnych buniek. – - – predpona označujúca 1. staršie označenie druhého atómu uhlíka v reťazci, na kt. sa pripája hlavná funkčná skupina, napr. kys. -hydroxymaslová správ. kys. 3-hydroxymaslová; 2. špecifická rotácia opticky aktívnej látky, napr. -D-glukóza; 3. orientácia exocyklického atómu al. skupiny, napr. cholest-5-en-3--ol; 4. plazmatický proteín, kt. migruje v elektroforéze v pruhu - lipoproteín; 5. člen série príbuzných chem. látok, najmä série stereoizomérov, izomérov, polymérov al. alotroických foriem, napr. -karotén; 6. -lúč. B2 – staršie označenie pre CD21. B4 – staršie označenie pre CD 19. B19 virus – ľudský parvovírus z čeľade Parvoviridae, značne rozšírený, vyvolávajúci obvykle inaparentné infekcie. -

974-Form.Pdf
California Association for Medical Laboratory Technology Distance Learning Program ANAEROBIC BACTERIOLOGY FOR THE CLINICAL LABORATORY by James I. Mangels, MA, CLS, MT(ASCP) Consultant Microbiology Consulting Services Santa Rosa, CA Course Number: DL-974 3.0 CE/Contact Hour Level of Difficulty: Intermediate © California Association for Medical Laboratory Technology. Permission to reprint any part of these materials, other than for credit from CAMLT, must be obtained in writing from the CAMLT Executive Office. CAMLT is approved by the California Department of Health Services as a CA CLS Accrediting Agency (#0021) and this course is is approved by ASCLS for the P.A.C.E. ® Program (#519) 1895 Mowry Ave, Suite 112 Fremont, CA 94538-1766 Phone: 510-792-4441 FAX: 510-792-3045 Notification of Distance Learning Deadline All continuing education units required to renew your license must be earned no later than the expiration date printed on your license. If some of your units are made up of Distance Learning courses, please allow yourself enough time to retake the test in the event you do not pass on the first attempt. CAMLT urges you to earn your CE units early!. CAMLT Distance Learning Course # DL-974 1 © California Association for Medical Laboratory Technology Outline A. Introduction B. What are anaerobic bacteria? Concepts of anaerobic bacteriology C. Why do we need to identify anaerobes? D. Normal indigenous anaerobic flora; the incidence of anaerobes at various body sites E. Anaerobic infections; most common anaerobic infections F. Specimen collection and transport; acceptance and rejection criteria G. Processing of clinical specimens 1. Microscopic examination 2. -

Identification and Antimicrobial Susceptibility Testing of Anaerobic
antibiotics Review Identification and Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Rubik’s Cube of Clinical Microbiology? Márió Gajdács 1,*, Gabriella Spengler 1 and Edit Urbán 2 1 Department of Medical Microbiology and Immunobiology, Faculty of Medicine, University of Szeged, 6720 Szeged, Hungary; [email protected] 2 Institute of Clinical Microbiology, Faculty of Medicine, University of Szeged, 6725 Szeged, Hungary; [email protected] * Correspondence: [email protected]; Tel.: +36-62-342-843 Academic Editor: Leonard Amaral Received: 28 September 2017; Accepted: 3 November 2017; Published: 7 November 2017 Abstract: Anaerobic bacteria have pivotal roles in the microbiota of humans and they are significant infectious agents involved in many pathological processes, both in immunocompetent and immunocompromised individuals. Their isolation, cultivation and correct identification differs significantly from the workup of aerobic species, although the use of new technologies (e.g., matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, whole genome sequencing) changed anaerobic diagnostics dramatically. In the past, antimicrobial susceptibility of these microorganisms showed predictable patterns and empirical therapy could be safely administered but recently a steady and clear increase in the resistance for several important drugs (β-lactams, clindamycin) has been observed worldwide. For this reason, antimicrobial susceptibility testing of anaerobic isolates for surveillance -

(BCID) Results Are “Not Detected”
Interpretation of Positive Blood Cultures When PCR Blood Culture Identification (BCID) Results are “Not Detected” Nebraska Medicine currently uses a multi-plex PCR-based blood culture identification (BCID) system that is able to identify 19 potential pathogens growing in blood culture. BCID generally detects over 90% of the most common causative agents in bloodstream infections; however, when microbes not included on the panel are present in a blood culture, it returns a result of “Not Detected.” This document aims to provide guidance in these scenarios supported by data collected at Nebraska Medicine from January 2018 to August 2019. Table 1: Recommendations for treatment of patients with blood cultures growing organisms not detected on BCID Gram Stain/Preliminary Likely Organism (% total BCID negative)* Recommended Treatment Culture Result Gram-positive: Aerobe Micrococcus sp. (18.1%) (most can also grow in Coagulase-negative Staphylococcus (9.3%) None anaerobic bottles) Diphtheroids (7%) None Peptostreptococcus sp. (4.4%) If therapy is desired: Anaerobe bottle only Lactobacillus sp. (2.6%) Metronidazole 500 mg PO q8h Clostridium sp. (2.6%) OR Penicillin G 4 million units IV q4h Gram-negative: Aerobe Acinetobacter sp. (1.8%) (most can also grow in Stenotrophomonas maltophilia (1.6%) Levofloxacin 750 mg IV/PO q24h anaerobic bottles) Pseudomonas fluorescens-putida group (1%) Bacteroides fragilis group (9.3%) Anaerobe bottle only Metronidazole 500 mg IV/PO q8h Fusobacterium sp. (4.7%) *A full list of isolated organisms can be found below in Table 2 Orange text = Cocci, Blue text = Bacilli (rods) Gram-Positives When BCID results as “Not Detected” but there is microbial growth, the organism is most frequently gram-positive (71%). -

Extraintestinal Clostridioides Difficile Infections
antibiotics Article Extraintestinal Clostridioides difficile Infections: Epidemiology in a University Hospital in Hungary and Review of the Literature Edit Urbán 1,*, Gabriella Terhes 2 and Márió Gajdács 3 1 Department of Public Health, Faculty of Medicine, University of Szeged, Dóm tér 10., 6720 Szeged, Hungary 2 Institute of Clinical Microbiology, Faculty of Medicine, University of Szeged, Semmelweis utca 6., 6725 Szeged, Hungary; [email protected] 3 Department of Pharmacodynamics and Biopharmacy, Faculty of Pharmacy, University of Szeged, Eötvös utca 6., 6720 Szeged, Hungary; [email protected] * Correspondence: [email protected]; Tel.: +36-62-342-861 Received: 8 December 2019; Accepted: 31 December 2019; Published: 2 January 2020 Abstract: Extraintestinal manifestations of Clostridioides difficile infections (CDIs) are very uncommon, and according to the literature, poor outcomes and a high mortality have been observed among affected individuals. The objective of this study was to investigate the incidence rate of extraintestinal infections caused by C. difficile (ECD) in a tertiary-care university hospital in Hungary. During a 10-year study period, the microbiology laboratory isolated 4129 individual strains of C. difficile; among these, the majority were either from diarrheal fecal samples or from colonic material and only n = 24 (0.58%) were from extraintestinal sources. The 24 extraintestinal C. difficile isolates were recovered from 22 patients (female-to-male ratio: 1, average age: 55.4 years). The isolates in n = 8 patients were obtained from abdominal infections, e.g., appendicitis, rectal abscess or Crohn’s disease. These extraintestinal cases occurred without concomitant diarrhea. In all, but two cases C. -
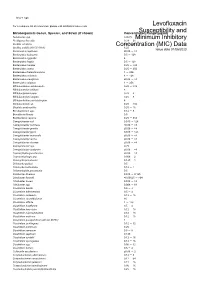
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 - -

A Case of Necrotizing Fasciitis Caused by Finegoldia Magna in a Patient with Type 2 Diabetes Mellitus
Le Infezioni in Medicina, n. 4, 359-363, 2018 CASE REPORTS 359 A case of necrotizing fasciitis caused by Finegoldia magna in a patient with type 2 diabetes mellitus Margherita Scapaticci1, Sabina Marchetto2, Andrea Nardi2, Maira Zoppelletto3, Andrea Bartolini3 1Laboratory Medicine Department, San Camillo Hospital, Treviso, Italy; 2Diabetic Foot Surgery Department, San Camillo Hospital, Treviso, Italy; 3Laboratory Medicine, San Bassiano Hospital, AULSS 7 Pedemontana, Bassano del Grappa, Italy SUMMARY Diabetes mellitus is one of the serious conditions the appropriate antibacterial agents. Hence the key associated with necrotizing fasciitis, a severe bac- to successful management is an early and accurate terial skin infection that spreads quickly and is diagnosis. We report a case of necrotizing fasciitis characterized by extensive necrosis of the deep and caused by Finegoldia magna in a patient with type 2 superficial fascia resulting in devascularization diabetes mellitus. and necrosis of the associated tissues. In addition to debridement and aggressive surgery procedures, Keywords: necrotizing fasciitis, type 2 diabetes mellitus, the effectiveness of therapy depends on choosing diabetic foot ulcers, anaerobes, GPACs. n INTRODUCTION lesion caused by a trauma, frequently trivial [4]. Symptoms include red or purple skin in the affect- iabetic foot ulcers are one of the main caus- ed area, severe pain, fever, and vomiting [5]. The Des of hospitalization and the major cause of major causative organisms include Streptococcus morbidity in individuals suffering from diabetes pyogenes, Staphylococcus aureus, anaerobic bacteria and, if not properly treated, can need amputation. and intestinal flora [4,6]. Diabetic patients may be The effectiveness of therapy depends on choos- predisposed to necrotizing fasciitis by the tissue hy- ing the appropriate antibacterial agents, and for poxia caused by arteriosclerosis and the immuno- this reason the key to a successful management is deficiency associated with poor glycemic control. -

Clinically Relevant Gram- Positive Anaerobic Cocci (GPAC), New Names and Their Antibiotic Susceptibility
Clinically relevant Gram- positive anaerobic cocci (GPAC), new names and their antibiotic susceptibility © by author ESCMID Online Lecture Library Linda Veloo Contents Introduction Gram-positive anaerobic cocci Peptostreptococcus sp. (formerly) New species and taxonomy Identification : - Phenotypic - Vitek ANC© bycard author - Fluorescent in situ hybridisation (FISH) - MALDI-TOF MS ESCMID Online Lecture Library Antibiotic resistance Virulence F. magna Introduction Gram-positive anaerobic cocci (GPAC): Peptococcus P.niger Seldomly isolated from clinical material Ruminococcus Faecel microbiota Seldomly ©isolated by fromauthor clinical material Insufficient culture methods? ESCMIDCoprococcus Online Lecture Library Isolated from faecal samples Gram positive anaerobic cocci, D.A. Murdoch Clinical Microbiology reviews, 1998 Introduction Atopobium Faecal microbiota and clinical material Sarcina Soil, abdominal samples, faecal samples from vegetarians and patients with gastro-intestinal abnormalities Unique cell© morphology: by author Groups of 8 cells which might produce spores ESCMID Online Lecture Library Peptostreptococcus The genus Peptostreptococcus contained 13 species. Clinically most relevant (literature): P. anaerobius P. asaccharolyticus P. magnus P. micros © by author ESCMID Online Lecture Library New species, changed taxonomy (in chronological order) In 1997 3 new species were added to the genus Peptostreptococcus P. harei P. ivorii P. octavius Clinical relevance unknown© by author ESCMID Online Lecture Library Description of Three New Species of the Genus Peptostreptococcus from Human Clinical Specimens: P. harei sp. nov., P. ivorii sp. nov., and P. octavius sp. nov., D. A. Murdoch et al., Int. J. Syst. Bact. 1997 New taxonomy Reclassification of Peptostreptococcus magnus (Prevot 1933) Holdeman and Moore 1972 as Finegoldia magna comb. nov. and Peptostreptococcus micros (Prevot 1933) Smith 1957 as Micromonas micros comb. -

Streptococcus Agalactiae, Arcanobacterium Haemolyticum과 Finegoldia Magna에 의한 괴사성 근막염 1예
대한진단검사의학회지 제 28권제3호 2008 � 증례∙임상미생물학 � Korean J Lab Med 2008;28:191-5 DOI 10.3343/kjlm.2008.28.3.191 개에게 물린 당뇨환자의 Streptococcus agalactiae, Arcanobacterium haemolyticum과 Finegoldia magna에 의한 괴사성 근막염 1예 이성실1∙노경호2∙김창기1∙용동은1,3∙최준용 4∙이진우∙이경원1,3∙정윤섭1 연세대학교 의과대학 진단검사의학교실 세균내성연구소1, BK21 의과학사업단 3, 내과학교실4, 정형외과학교실5, 고려대학교 의과대학 진단검사의학교실2 A Case of Necrotizing Fasciitis Due to Streptococcus agalactiae, Arcanobacterium haemolyticum, and Finegoldia magna in a Dog-bitten Patient with Diabetes Sungsil Lee, M.D.1, Kyoung Ho Roh, M.D.2, Chang Ki Kim, M.D.1, Dongeun Yong, M.D.1,3, Jun Yong Choi, M.D.4, Jin Woo Lee, M.D.5, Kyungwon Lee, M.D.1,3, and Yunsop Chong, Ph.D.1 Departments of Laboratory Medicine and Research Institute of Bacterial Resistance1; BK21 Project for Medical Sciences3, Internal Medicine4, and Orthopedic Surgery5, Yonsei University College of Medicine, Seoul; Department of Laboratory Medicine2, Korea University College of Medicine, Seoul, Korea We report a case of necrotizing fasciitis involving Streptococcus agalactiae, Arcanobacterium haemolyticum, and Finegoldia magna in a 36-yr-old female diabetic patient, which started after a minor dog bite to the toe of the patient. This case suggested that a trivial infection after a minor dog bite in an immunocompromised patient such as diabetes patient could result in a significant compli- cation such as necrotizing fasciitis. The life-threatening infection was cured by timely above-the-knee amputation, as well as penicillin G and clindamycin therapy. (Korean J Lab Med 2008;28:191-5) Key Words : Necrotizing fasciitis, Streptococcus agalactiae, Arcanobacterium haemolyticum, Finegoldia magna, Diabetic patient INTRODUCTION tions[1-3].