Detoxification of Arsenic and Chromium Through Chelators and Enzymes: an In-Silico Approach
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Effects of Calcium Chelation on Digitalis-Induced Cardiac Arrhythmias by Paul Szekely and N
Br Heart J: first published as 10.1136/hrt.25.5.589 on 1 September 1963. Downloaded from Brit. Heart J., 1963, 25, 589. EFFECTS OF CALCIUM CHELATION ON DIGITALIS-INDUCED CARDIAC ARRHYTHMIAS BY PAUL SZEKELY AND N. A. WYNNE From the Cardiovascular Department, Newcastle General Hospital, and the Department ofPhysiology, King's College, University of Durham Received November 19, 1962 Several studies have already shown that cardiac arrhythmias caused by digitalis can be abolished by the induction of hypocalcaemia (Page and Real, 1955; Smith and Grinnell, 1955; Gubner and Kallman, 1957; Kabakow and Brothers, 1958; Surawicz et al., 1959; Cohen et al., 1959; Rosenbaum, Mason, and Seven, 1960; Surawicz, 1960; Soffer, Toribara, and Sayman, 1961). The rationale of inducing hypocalcemia as an anti-arrhythmic measure is based on experimental and clinical obser- vations relating to the direct myocardial action of calcium and to the interaction between calcium, potassium, and digitalis. Low calcium concentration decreases myocardial irritability (Brooks et al., 1955) and it also increases the intracellular potassium concentration (Rosenbaum et al., 1960). There is also a synergistic action between calcium and digitalis, which was observed in the presence of digitalis intoxication (Nalbandian et al., 1957). The present study was undertaken copyright. in order to assess the value of induced hypocalcemia in the management of cardiac arrhythmias caused by digitalis. MATERIAL AND METHODS Forty-eight experiments were carried out under general aneesthesia on 46 cats and 2 dogs. Cardiac arrhythmia was induced by the intravenous administration of tincture of digitalis as previously described http://heart.bmj.com/ (Szekely and Wynne, 1951). -

Safety Data Sheet
SAFETY DATA SHEET Creation Date 22-Sep-2009 Revision Date 24-Jun-2020 Revision Number 4 1. Identification Product Name Diethylenetriaminepentaacetic acid Cat No. : AC114320000; AC114320010; AC114320050; AC114322500 CAS-No 67-43-6 Synonyms (Carboxymethylimino)bis(ethylenenitrilo)tetraacetic acid; DTPA; Pentetic acid Recommended Use Laboratory chemicals. Uses advised against Food, drug, pesticide or biocidal product use. Details of the supplier of the safety data sheet Company Fisher Scientific Company Acros Organics One Reagent Lane One Reagent Lane Fair Lawn, NJ 07410 Fair Lawn, NJ 07410 Tel: (201) 796-7100 Emergency Telephone Number For information US call: 001-800-ACROS-01 / Europe call: +32 14 57 52 11 Emergency Number US:001-201-796-7100 / Europe: +32 14 57 52 99 CHEMTREC Tel. No.US:001-800-424-9300 / Europe:001-703-527-3887 2. Hazard(s) identification Classification This chemical is considered hazardous by the 2012 OSHA Hazard Communication Standard (29 CFR 1910.1200) Acute Inhalation Toxicity - Dusts and Mists Category 4 Serious Eye Damage/Eye Irritation Category 2 Reproductive Toxicity Category 2 Specific target organ toxicity - (repeated exposure) Category 2 Inhalation Label Elements Signal Word Warning Hazard Statements Causes serious eye irritation Harmful if inhaled Suspected of damaging fertility or the unborn child May cause damage to organs through prolonged or repeated exposure ______________________________________________________________________________________________ Page 1 / 7 Diethylenetriaminepentaacetic acid Revision -

New Brunswick Drug Plans Formulary
New Brunswick Drug Plans Formulary August 2019 Administered by Medavie Blue Cross on Behalf of the Government of New Brunswick TABLE OF CONTENTS Page Introduction.............................................................................................................................................I New Brunswick Drug Plans....................................................................................................................II Exclusions............................................................................................................................................IV Legend..................................................................................................................................................V Anatomical Therapeutic Chemical (ATC) Classification of Drugs A Alimentary Tract and Metabolism 1 B Blood and Blood Forming Organs 23 C Cardiovascular System 31 D Dermatologicals 81 G Genito Urinary System and Sex Hormones 89 H Systemic Hormonal Preparations excluding Sex Hormones 100 J Antiinfectives for Systemic Use 107 L Antineoplastic and Immunomodulating Agents 129 M Musculo-Skeletal System 147 N Nervous System 156 P Antiparasitic Products, Insecticides and Repellants 223 R Respiratory System 225 S Sensory Organs 234 V Various 240 Appendices I-A Abbreviations of Dosage forms.....................................................................A - 1 I-B Abbreviations of Routes................................................................................A - 4 I-C Abbreviations of Units...................................................................................A -

Review of Succimer for Treatment of Lead Poisoning
Review of Succimer for treatment of lead poisoning Glyn N Volans MD, BSc, FRCP. Department of Clinical Pharmacology, School of Medicine at Guy's, King's College & St Thomas' Hospitals, St Thomas' Hospital, London, UK Lakshman Karalliedde MB BS, DA, FRCA Consultant Medical Toxicologist, CHaPD (London), Health Protection Agency UK, Visiting Senior Lecturer, Division of Public Health Sciences, King's College Medical School, King's College , London Senior Research Collaborator, South Asian Clinical Toxicology Research Collaboration, Faculty of Medicine, Peradeniya, Sri Lanka. Heather M Wiseman BSc MSc Medical Toxicology Information Services, Guy’s and St Thomas’ NHS Foundation Trust, London SE1 9RT, UK. Contact details: Heather Wiseman Medical Toxicology Information Services Guy’s & St Thomas’ NHS Foundation Trust Mary Sheridan House Guy’s Hospital Great Maze Pond London SE1 9RT Tel 020 7188 7188 extn 51699 or 020 7188 0600 (admin office) Date 10th March 2010 succimer V 29 Nov 10.doc last saved: 29-Nov-10 11:30 Page 1 of 50 CONTENTS 1 Summary 2. Name of the focal point in WHO submitting or supporting the application 3. Name of the organization(s) consulted and/or supporting the application 4. International Nonproprietary Name (INN, generic name) of the medicine 5. Formulation proposed for inclusion 6. International availability 7. Whether listing is requested as an individual medicine or as an example of a therapeutic group 8. Public health relevance 8.1 Epidemiological information on burden of disease due to lead poisoning 8.2 Assessment of current use 8.2.1 Treatment of children with lead poisoning 8.2.2 Other indications 9. -

Chelation Therapy
Medical Policy Chelation Therapy Table of Contents • Policy: Commercial • Coding Information • Information Pertaining to All Policies • Policy: Medicare • Description • References • Authorization Information • Policy History Policy Number: 122 BCBSA Reference Number: 8.01.02 NCD/LCD: N/A Related Policies None Policy Commercial Members: Managed Care (HMO and POS), PPO, and Indemnity Medicare HMO BlueSM and Medicare PPO BlueSM Members Chelation therapy in the treatment of the following conditions is MEDICALLY NECESSARY: • Extreme conditions of metal toxicity • Treatment of chronic iron overload due to blood transfusions (transfusional hemosiderosis) or due to nontransfusion-dependent thalassemia (NTDT) • Wilson's disease (hepatolenticular degeneration), or • Lead poisoning. Chelation therapy in the treatment of the following conditions is MEDICALLY NECESSARY if other modalities have failed: • Control of ventricular arrhythmias or heart block associated with digitalis toxicity • Emergency treatment of hypercalcemia. NaEDTA as chelation therapy is considered NOT MEDICALLY NECESSARY. Off-label applications of chelation therapy are considered INVESTIGATIONAL, including, but not limited to: • Alzheimer’s disease • Arthritis (includes rheumatoid arthritis) • Atherosclerosis, (e.g., coronary artery disease, secondary prevention in patients with myocardial infarction, or peripheral vascular disease) • Autism • Diabetes • Multiple sclerosis. 1 Prior Authorization Information Inpatient • For services described in this policy, precertification/preauthorization IS REQUIRED for all products if the procedure is performed inpatient. Outpatient • For services described in this policy, see below for products where prior authorization might be required if the procedure is performed outpatient. Outpatient Commercial Managed Care (HMO and POS) Prior authorization is not required. Commercial PPO and Indemnity Prior authorization is not required. Medicare HMO BlueSM Prior authorization is not required. -

Post-Injury Calcium Chelation Rescues Skeletal Muscle Regeneration in Mice Matthew .D Magda University of Connecticut - Storrs, [email protected]
University of Connecticut OpenCommons@UConn Honors Scholar Theses Honors Scholar Program Summer 8-1-2013 Post-Injury Calcium Chelation Rescues Skeletal Muscle Regeneration in Mice Matthew .D Magda University of Connecticut - Storrs, [email protected] Follow this and additional works at: https://opencommons.uconn.edu/srhonors_theses Part of the Cell Biology Commons, and the Molecular Biology Commons Recommended Citation Magda, Matthew D., "Post-Injury Calcium Chelation Rescues Skeletal Muscle Regeneration in Mice" (2013). Honors Scholar Theses. 321. https://opencommons.uconn.edu/srhonors_theses/321 1 Post-Injury Calcium Chelation Rescues Skeletal Muscle Regeneration in Mice Honors Thesis Matthew Magda Research Advisor: Dr. Morgan Carlson Honors Advisor: Dr. Kenneth Noll August 2013 2 Abstract: Antibiotics, surgery and organ transplants have pushed average lifespans towards the upper limits of the human body. Drastically reduced morbidity from infection, toxins and traumatic injury have allowed ever greater portions of the populace can reach eighty or ninety years old before dying of old age. Despite the increased role of aging as a source of morbidity, many aspects of aging are poorly characterized. Sarcopenia, progressive muscle loss, and loss of adult myogenic potential, the ability to produce new muscle tissue from adult stem cell sources, are key causes of decreased mobility and strength in aged individuals. If more youthful muscle quality could be restored in old patients they would experience greatly improved quality of life and perhaps even longer lifespans. Satellite cell populations are known to decline sharply by 6-7th decade of life but traditional treatments for sarcopenia, namely exercise intervention, have been shown to exacerbate the degeneration in aged such patients. -

WO 2014/195872 Al 11 December 2014 (11.12.2014) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2014/195872 Al 11 December 2014 (11.12.2014) P O P C T (51) International Patent Classification: (74) Agents: CHOTIA, Meenakshi et al; K&S Partners | Intel A 25/12 (2006.01) A61K 8/11 (2006.01) lectual Property Attorneys, 4121/B, 6th Cross, 19A Main, A 25/34 (2006.01) A61K 8/49 (2006.01) HAL II Stage (Extension), Bangalore 560038 (IN). A01N 37/06 (2006.01) A61Q 5/00 (2006.01) (81) Designated States (unless otherwise indicated, for every A O 43/12 (2006.01) A61K 31/44 (2006.01) kind of national protection available): AE, AG, AL, AM, AO 43/40 (2006.01) A61Q 19/00 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, A01N 57/12 (2006.01) A61K 9/00 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, AOm 59/16 (2006.01) A61K 31/496 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (21) International Application Number: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, PCT/IB20 14/06 1925 KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (22) International Filing Date: OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, 3 June 2014 (03.06.2014) SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (25) Filing Language: English TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Chelation Therapy
Corporate Medical Policy Chelation Therapy File Name: chelation_therapy Origination: 12/1995 Last CAP Review: 2/2021 Next CAP Review: 2/2022 Last Review: 2/2021 Description of Procedure or Service Chelation therapy is an established treatment for the removal of metal toxins by converting them to a chemically inert form that can be excreted in the urine. Chelation therapy comprises intravenous or oral administration of chelating agents that remove metal ions such as lead, aluminum, mercury, arsenic, zinc, iron, copper, and calcium from the body. Specific chelating agents are used for particular heavy metal toxicities. For example, desferroxamine (not Food and Drug Administration [FDA] approved) is used for patients with iron toxicity, and calcium-ethylenediaminetetraacetic acid (EDTA) is used for patients with lead poisoning. Note that disodium-EDTA is not recommended for acute lead poisoning due to the increased risk of death from hypocalcemia. Another class of chelating agents, called metal protein attenuating compounds (MPACs), is under investigation for the treatment of Alzheimer’s disease, which is associated with the disequilibrium of cerebral metals. Unlike traditional systemic chelators that bind and remove metals from tissues systemically, MPACs have subtle effects on metal homeostasis and abnormal metal interactions. In animal models of Alzheimer’s disease, they promote the solubilization and clearance of β-amyloid protein by binding to its metal-ion complex and also inhibit redox reactions that generate neurotoxic free radicals. MPACs therefore interrupt two putative pathogenic processes of Alzheimer’s disease. However, no MPACs have received FDA approval for treating Alzheimer’s disease. Chelation therapy has also been investigated as a treatment for other indications including atherosclerosis and autism spectrum disorder. -

Chemical Compatibility Guide
Chemical Compatibility Guide Guide Applicable to the Following: PIG Portable Spill Containment Pool Guide Information This report is offered as a guide and was developed from information which, to the best of New Pig’s knowledge, was reliable and accurate. Due to variables and conditions of application beyond New Pig’s control, none of the data shown in this guide is to be construed as a guarantee, expressed or implied. New Pig assumes no responsibility, obligation, or liability in conjunction with the use or misuse of the information. PIG Spill Containment Pools are constructed from PVC-coated polyester fabric. The chemical resistance guide that follows shows the chemical resistance for the PVC layer only. This guide has been compiled to provide the user with general chemical resistance information. It does not reflect actual product testing. Ratings / Key or Ratings – Chemical Effect 1. Satisfactory to 72°F (22°C) 2. Satisfactory to 120°F (48°C) A = Excellent D = Severe Effect, not recommended for ANY use. B = Good — Minor Effect, slight corrosion or discoloration. N/A = Information not available. C = Fair — Moderate Effect, not recommended for continuous use. Softening, loss of strength, swelling may occur. Due to variables and conditions beyond our control, New Pig cannot guarantee that this product(s) will work to your satisfaction. To ensure effectiveness and your safety, we recommend that you conduct compatibility and absorption testing of your chemicals with this product prior to purchase. For additional questions or information, -

Chelating Drug Therapy: an Update
Open Access Austin Journal of Genetics and Genomic Research Review Article Chelating Drug Therapy: An Update Vijay Kumar1, Ashok Kumar2*, Sandeep Kumar Singh1, Manoj Kumar3, Surendra Kumar2, Dinesh Abstract 4 5 Kumar and Ragni Singh Purpose: To study the clinical effects of metal toxicity and current 1Department of Neurology, SGPGIMS, India recommendations for management, including chelation therapy, are reviewed. 2Department of Medical Genetics, SGPGIMS, India 3Department of Microbiology, SGPGIMS, India Summary: Metals are essential to many biological processes, but excess 4Department of Chemistry, Dr. R.M.L. Avadh University, of it becomes hazardous to life. These are necessary for cell growth, electron India transport chain, several enzymatic activities and response of immune systems. 5Bheem Rao Ambedkar Bihar University, India They also serve as a cofactor for several enzymes. Chelation therapy is used for clinical management of the excess of metal. However, each metal requires *Corresponding author: Ashok Kumar, Department a specific chelation agent. A chelate is a compound form between metal and a of Medical Genetics, Sanjay Gandhi Post Graduate compound that contains two or more potential ligands. A promising Fe chelator Institute of Medical Sciences, Lucknow, India is Desferrioxamine (Desferal). Penicillamine and Trientine are uses for copper Received: March 12, 2015; Accepted: April 24, 2015; chelation. Meso-2,3-Dimercaptosuccinic Acid (DMSA) and 2,3-Dimercapto- Published: April 27, 2015 Propanesulphonate (DMPS) can be used as effective chelator of mercury. Dimercaprol, edetate calcium disodium, and succimer are the three agents primarily used for chelation of lead. Conclusion: Metal toxicity remains a significant public health concern. Elimination of elevated metal ions can be achieved by proper chelation agents. -

Changes in Tissue Gadolinium Biodistribution Measured in an Animal Model Exposed to Four Chelating Agents
Japanese Journal of Radiology (2019) 37:458–465 https://doi.org/10.1007/s11604-019-00835-1 ORIGINAL ARTICLE Changes in tissue gadolinium biodistribution measured in an animal model exposed to four chelating agents Türker Acar1,6 · Egemen Kaya2 · Mustafa Deniz Yoruk3 · Neslihan Duzenli4 · Recep Selim Senturk4 · Cenk Can4 · Lokman Ozturk3 · Canberk Tomruk5 · Yigit Uyanikgil5 · Frank J. Rybicki6 Received: 17 December 2018 / Accepted: 22 March 2019 / Published online: 30 March 2019 © Japan Radiological Society 2019 Abstract Purpose This study investigated the potential to reduce gadolinium levels in rodents after repetitive IV Gadodiamide admin- istration using several chelating agents. Materials and methods The following six groups of rats were studied. Group 1: Control; Group 2: Gadodiamide only; Group 3: Meso-2,3-Dimercaptosuccinic acid (DMSA) + Gadodiamide; Group 4: N-Acetyl-L-cysteine (NAC) + Gadodiamide; Group 5: Coriandrum sativum extract + Gadodiamide; and Group 6: Deferoxamine + Gadodiamide. Brain, kidney, and blood samples were evaluated via inductively coupled plasma mass spectrometry. The brain was also evaluated histologically. Results Kidney gadolinium levels in Groups 4 and 5 were approximately double that of Group 2 (p = 0.033 for each). There was almost no calcifcation in rat hippocampus for Group 4 rodents when compared with Groups 2, 3, 5 and 6. Conclusion Our preliminary study shows that excretion to the kidney has a higher propensity in NAC and Coriandrum sati- vum groups. It may be possible to change the distribution of gadolinium by administrating several agents. NAC may lower Gadodiamide-induced mineralization in rat hippocampus. Keywords Gadolinium deposition · Dimercaptosuccinic acid · N-Acetyl-L-cysteine · Coriandrum sativum · Deferoxamine Introduction However, the safety profle of gadolinium-based agents [1] was questioned after the discovery of nephrogenic sys- Gadolinium-based contrast agents (GBCA) have been safely temic fbrosis [2], a scleroderma-like disease characterized used in diagnostic radiology since the 1980s. -
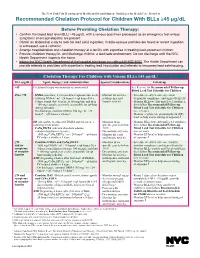
Recommended Chelation Protocol for Children with Blls ≥45 Μg/Dl
The New York City Department of Health and Mental Hygiene Guidelines for Health Care Providers Recommended Chelation Protocol for Children With BLLs ≥45 μg/dL Before Providing Chelation Therapy: • Confirm the blood lead level (BLL) ≥45 μg/dL with a venous specimen processed as an emergency test unless symptoms of encephalopathy are present. • Obtain an abdominal x-ray to look for lead solid ingestion; if radio-opaque particles are found or recent ingestion is witnessed, use a cathartic. • Arrange hospitalization and chelation therapy at a facility with expertise in treating lead-poisoned children. • Provide chelation therapy in, and discharge child to, a lead-safe environment. Do not discharge until the NYC Health Department inspects the home. • Inform the NYC Health Department of the hospital admission by calling 646-632-6002. The Health Department can provide referrals to providers with expertise in treating lead intoxication and referrals to temporary lead-safe housing. Chelation Therapy For Children with Venous BLLs ≥45 μg/dL1 BLL (μg/dL) Agent, Dosage,* and Administration Special Considerations Follow-up <45 Chelation therapy not routinely recommended See Reverse for Recommended Follow-up Blood Lead Test Schedule for Children 45 to <70 • DMSA (succimer, 2,3-meso-dimercaptosuccinic acid) • Monitor for anemia, • Schedule weekly health care visits • 1050 mg DMSA / m2 / 24 hours* ÷ q8 hours PO x neutropenia, and to monitor compliance and signs of toxicity. 5 days; round dose to nearest 100 mg/day, and then hepatic toxicity. • Monitor BLLs weekly until level stabilizes, ÷ 100-mg capsules as evenly as possible for q8-hour then follow Recommended Follow-up dosing schedule.