Title of Dissertation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Supplementary Methods
Supplementary methods Human lung tissues and tissue microarray (TMA) All human tissues were obtained from the Lung Cancer Specialized Program of Research Excellence (SPORE) Tissue Bank at the M.D. Anderson Cancer Center (Houston, TX). A collection of 26 lung adenocarcinomas and 24 non-tumoral paired tissues were snap-frozen and preserved in liquid nitrogen for total RNA extraction. For each tissue sample, the percentage of malignant tissue was calculated and the cellular composition of specimens was determined by histological examination (I.I.W.) following Hematoxylin-Eosin (H&E) staining. All malignant samples retained contained more than 50% tumor cells. Specimens resected from NSCLC stages I-IV patients who had no prior chemotherapy or radiotherapy were used for TMA analysis by immunohistochemistry. Patients who had smoked at least 100 cigarettes in their lifetime were defined as smokers. Samples were fixed in formalin, embedded in paraffin, stained with H&E, and reviewed by an experienced pathologist (I.I.W.). The 413 tissue specimens collected from 283 patients included 62 normal bronchial epithelia, 61 bronchial hyperplasias (Hyp), 15 squamous metaplasias (SqM), 9 squamous dysplasias (Dys), 26 carcinomas in situ (CIS), as well as 98 squamous cell carcinomas (SCC) and 141 adenocarcinomas. Normal bronchial epithelia, hyperplasia, squamous metaplasia, dysplasia, CIS, and SCC were considered to represent different steps in the development of SCCs. All tumors and lesions were classified according to the World Health Organization (WHO) 2004 criteria. The TMAs were prepared with a manual tissue arrayer (Advanced Tissue Arrayer ATA100, Chemicon International, Temecula, CA) using 1-mm-diameter cores in triplicate for tumors and 1.5 to 2-mm cores for normal epithelial and premalignant lesions. -

Efficient Entry to Both Enantiomers of Α-Halohydrins Employing Two
Electronic Supplementary Material (ESI) for RSC Advances. This journal is © The Royal Society of Chemistry 2015 Supporting information Enantioselectively bioreductive preparation of chiral halohydrins employing two newly identified stererocomplementary reductases Guo-chao Xu,1,2 Hui-lei Yu,1,* Yue-peng Shang,1 Jian-he Xu1 1 State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, and Shanghai Collaborative Innovation Center for Biomanufacturing Technology, Shanghai 200237, China. 2 The Key Laboratory of Industrial Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, China. *Corresponding authors. E-mail: [email protected]; [email protected]. Table S1 Typical sequence motifs of SDR and AKR superfamily found in DhCR and CgCR..........2 Table S2 The secondary structure elements motif in ‘classical’ SDR, ‘extended’ SDR and DhCR.3 Table S3 Steady-state kinetic constants of stererocomplementary DhCR and CgCR......................4 Table S4 Substrate specificities of stererocomplementary DhCR and CgCR. ...................................5 Table S5 Concentrations of NAD+ and NADP+ in the fresh wet cells and dry cells. ..........................6 Table S6 Optimization of DhCR and CgCR catalyzed asymmetric reduction of COBE....................7 Table S7 Comparison of the characteristics of reported enzymes that produce optically active CHBE. ............................................................................................................................................................8 -

Development of Potent and Selective Inhibitors of Aldo−Keto Reductase
Article pubs.acs.org/jmc Development of Potent and Selective Inhibitors of Aldo−Keto Reductase 1C3 (Type 5 17β-Hydroxysteroid Dehydrogenase) Based on N-Phenyl-Aminobenzoates and Their Structure−Activity Relationships † § ‡ § † † † ‡ Adegoke O. Adeniji, , Barry M. Twenter, , Michael C. Byrns, Yi Jin, Mo Chen, Jeffrey D. Winkler,*, † and Trevor M. Penning*, † Department of Pharmacology and Center of Excellence in Environmental Toxicology, Perelman School of Medicine, University of Pennsylvania, 130C John Morgan Building, 3620 Hamilton Walk, Philadelphia, Pennsylvania 19104-6084, United States ‡ Department of Chemistry, University of Pennsylvania, 231 South 34th Street, Philadelphia, Pennsylvania 19104, United States *S Supporting Information ABSTRACT: Aldo−keto reductase 1C3 (AKR1C3; type 5 17β-hydroxy- steroid dehydrogenase) is overexpressed in castration resistant prostate cancer (CRPC) and is implicated in the intratumoral biosynthesis of testosterone and 5α-dihydrotestosterone. Selective AKR1C3 inhibitors are required because compounds should not inhibit the highly related AKR1C1 and AKR1C2 isoforms which are involved in the inactivation of 5α-dihydrotestosterone. NSAIDs, N-phenylanthranilates in particular, are potent but nonselective AKR1C3 inhibitors. Using flufenamic acid, 2-{[3-(trifluoromethyl)phenyl]amino}- benzoic acid, as lead compound, five classes of structural analogues were synthesized and evaluated for AKR1C3 inhibitory potency and selectivity. Structure−activity relationship (SAR) studies revealed that a meta-carboxylic acid group relative to the amine conferred pronounced AKR1C3 selectivity without loss of potency, while electron withdrawing groups on the phenylamino B-ring were optimal for AKR1C3 inhibition. Lead compounds did not inhibit COX-1 or COX-2 but blocked the AKR1C3 mediated production of testosterone in LNCaP-AKR1C3 cells. These compounds offer promising leads toward new therapeutics for CRPC. -
Data Supplement
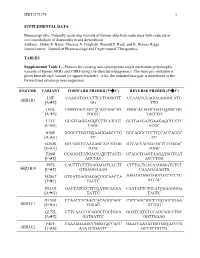
JPET#173179 1 SUPPLEMENTAL DATA Manuscript title: Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin Authors: Onkar S. Bains, Thomas A. Grigliatti, Ronald E. Reid, and K. Wayne Riggs Journal name: Journal of Pharmacology and Experimental Therapeutics TABLES Supplemental Table 1—Primers for creating non-synonymous single nucleotide polymorphic variants of human AKRs and CBR4 using site-directed mutagenesis. The base pair mutation is given beneath each variant [in square brackets]. Also, the mutated base pair is underlined in the forward and reverse primer sequences. ENZYME VARIANT FORWARD PRIMER (5'3') REVERSE PRIMER (5'3') I15F CAAGATGCCCTTCCTGGGGTT CCAACCCCAGGAAGGGCATC AKR1B1 [AT] GG TTG H42L CGGGTACCGCCTCATCGACTG GGGCACAGTCGATGAGGCGG [AT] TGCCC TACCCG L73V GCGTGAGGAGGTCTTCATCGT GCTGACGATGAAGACCTCCTC [CG] CAGC ACGC K90E GGGCCTGGTGGAAGGAGCCTG GGCAGGCTCCTTCCACCAGGC [AG] CC CC G204S GCCAGTCCAAAAGCATCGTGG GTCACCACGATGCTTTTGGAC [GA] TGAC TGGC T288I CCAGGATATGACCATCTTACTC GTAGCTGAGTAAGATGGTCAT [CT] AGCTAC ATCCTGG P87S CACTTTCTTTGAGAGATCCCTT CTTTCCTCACAAGGGATCTCT AKR1B10 [CT] GTGAGGAAAG CAAAGAAAGTG M286T GTGATGAGGAGACGGCAACCA GAGTATGGTTGCCGTCTCCTC [TC] TACTC ATCAC N313D GACTATCCCTTCGATGCAGAA CAATATTCTGCATCGAAGGGA [AG] TATTG TAGTC R170H CCAACTTCAACCACAGGCAGC CTCCAGCTGCCTGTGGTTGAA AKR1C1 [GA] TGGAG GTTGG Q172L CTTCAACCGCAGGCTGCTGGA GGATCATCTCCAGCAGCCTGC [AT] GATGATCC GGTTGAAG F46Y CAATAGAAGCCGGGTACCACC GAATCAATATGGTGGTACCCG AKR1C2 [TA] ATATTTGATTC GCTTCTATTG JPET#173179 -

Metabolic Enzyme/Protease
Inhibitors, Agonists, Screening Libraries www.MedChemExpress.com Metabolic Enzyme/Protease Metabolic pathways are enzyme-mediated biochemical reactions that lead to biosynthesis (anabolism) or breakdown (catabolism) of natural product small molecules within a cell or tissue. In each pathway, enzymes catalyze the conversion of substrates into structurally similar products. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation. Metabolism maintains the living state of the cells and the organism. Proteases are used throughout an organism for various metabolic processes. Proteases control a great variety of physiological processes that are critical for life, including the immune response, cell cycle, cell death, wound healing, food digestion, and protein and organelle recycling. On the basis of the type of the key amino acid in the active site of the protease and the mechanism of peptide bond cleavage, proteases can be classified into six groups: cysteine, serine, threonine, glutamic acid, aspartate proteases, as well as matrix metalloproteases. Proteases can not only activate proteins such as cytokines, or inactivate them such as numerous repair proteins during apoptosis, but also expose cryptic sites, such as occurs with β-secretase during amyloid precursor protein processing, shed various transmembrane proteins such as occurs with metalloproteases and cysteine proteases, or convert receptor agonists into antagonists and vice versa such as chemokine conversions carried out by metalloproteases, dipeptidyl peptidase IV and some cathepsins. In addition to the catalytic domains, a great number of proteases contain numerous additional domains or modules that substantially increase the complexity of their functions. -

BCAT1 Expression Associates with Ovarian Cancer Progression: Possible Implications in Altered Disease Metabolism
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 31 BCAT1 expression associates with ovarian cancer progression: possible implications in altered disease metabolism Zhi-Qiang Wang1,2, Adnen Faddaoui1,2, Magdalena Bachvarova2, Marie Plante2,3, Jean Gregoire2,3, Marie-Claude Renaud2,3, Alexandra Sebastianelli2,3, Chantal Guillemette4,5, Stéphane Gobeil1,4, Elizabeth Macdonald6, Barbara Vanderhyden6, Dimcho Bachvarov1,2 1Department of Molecular Medicine, Laval University, Québec PQ, Canada 2Centre de recherche du CHU de Québec, L’Hôtel-Dieu de Québec, Québec PQ, Canada 3Department of Obstetrics and Gynecology, Laval University, Québec PQ, Canada 4Centre de recherche du CHU de Québec, CHUL, Québec PQ, Canada 5Faculty of Pharmacy, Laval University, Québec PQ, Canada 6Department of Cellular and Molecular Medicine, University of Ottawa, Ottawa, ON, Canada Correspondence to: Dimcho Bachvarov, e-mail: [email protected] Keywords: BCAT1, ovarian cancer, cancer metabolism, metastasis, DNA hypomethylation Received: May 07, 2015 Accepted: August 28, 2015 Published: September 10, 2015 ABSTRACT Previously, we have identified the branched chain amino-acid transaminase 1 (BCAT1) gene as notably hypomethylated in low-malignant potential (LMP) and high-grade (HG) serous epithelial ovarian tumors, compared to normal ovarian tissues. Here we show that BCAT1 is strongly overexpressed in both LMP and HG serous epithelial ovarian tumors, which probably correlates with its hypomethylated status. Knockdown of the BCAT1 expression in epithelial ovarian cancer (EOC) cells led to sharp decrease of cell proliferation, migration and invasion and inhibited cell cycle progression. BCAT1 silencing was associated with the suppression of numerous genes and pathways known previously to be implicated in ovarian tumorigenesis, and the induction of some tumor suppressor genes (TSGs). -

Biostatistics Mining Associated Method Identifies AKR1B10
www.nature.com/scientificreports OPEN Biostatistics mining associated method identifes AKR1B10 enhancing hepatocellular Received: 26 February 2018 Accepted: 25 June 2018 carcinoma cell growth and Published: xx xx xxxx degenerated by miR-383-5p Junqing Wang 1,2,3, Yunyun Zhou4, Xiaochun Fei5, Xuehua Chen2,3 & Yongjun Chen1 Previous studies have reported that the aberrantly expressed AKR1B10 is associated with many cancer development, however the functional roles of AKR1B10 and its regulatory mechanisms in hepatocellular carcinoma (HCC) have been limited studied. In this project, we identifed AKR1B10 functional as an oncogene in HCC through tumor/normal human tissue comparison from both GEO microarray and TCGA RNAseq dataset. Further experimental validations from three HCC cell lines (SMMC-7721, HePG2 and HeP3B) also suggested the ontogenetic functions of AKR1B10 in HCC tumor growth. By knocking down AKR1B10 through shRNA in HCC HeP3B cells, we showed it signifcantly induced cell cycle arrest and inhibited cell growth. Interestingly, integrative analysis of TCGA RNAseq data and miRNA-seq data predicted that miR-383-5p, a novel post-transcriptional tumor suppressor, is negatively associated with AKR1B10 expression. To further investigate the role of miR-383-5p in regulating AKR1B10 in HCC, we performed Dual-luciferase reporter assay experiments. Results showed that miR-383-5p is an upstream modulator targeting AKR1B10 in the post-transcriptional stage. Thus, we report AKR1B10 modulated regulated by miR-383-5p, promotes HCC tumor progress, and could be potentially a therapeutic target for precision medicine in HCC. World widely, hepatocellular carcinoma (HCC) presents a high incidence rate among human malignancies, rank- ing a ffh of morbidity and second of malignancy-related mortality1,2. -

AKR1C1 Controls Cisplatin-Resistance in Head and Neck Squamous Cell
Chang et al. Journal of Experimental & Clinical Cancer Research (2019) 38:245 https://doi.org/10.1186/s13046-019-1256-2 RESEARCH Open Access AKR1C1 controls cisplatin-resistance in head and neck squamous cell carcinoma through cross-talk with the STAT1/3 signaling pathway Wei-Min Chang1, Yu-Chan Chang1, Yi-Chieh Yang1, Sze-Kwan Lin2, Peter Mu-Hsin Chang3,4*† and Michael Hsiao1,5*† Abstract Background: Cisplatin is the first-line chemotherapy used against most upper aerodigestive tract carcinomas. In head and neck cancer, sensitivity to cisplatin remains the key issue in treatment response and outcome. Genetic heterogeneity and aberrant gene expression may be the intrinsic factors that cause primary cisplatin-resistance. Methods: Combination of the HNSCC gene expression data and the cisplatin sensitivity results from public database. We found that aldo-keto reductase family 1 member C1 (AKR1C1) may be associated with cisplatin sensitivity in HNSCC treatment of naïve cells. We examined the AKR1C1 expression and its correlation with cisplatin IC50 and prognosis in patients. The in vitro and in vivo AKR1C1 functions in cisplatin-resistance through overexpression or knockdown assays, respectively. cDNA microarrays were used to identify the upstream regulators that modulate AKR1C1-induced signaling in HNSCC. Finally, we used the cigarette metabolites to promote AKR1C1 expression and ruxolitinib to overcome AKR1C1-induced cisplatin-resistance. Results: AKR1C1 positively correlates to cisplatin-resistance in HNSCC cells. AKR1C1 is a poor prognostic factor for recurrence and death of HNSCC patients. Silencing of AKR1C1 not only reduced in vitro IC50 but also increased in vivo cisplatin responses and vise versa in overexpression cells. -

Aldo-Keto Reductase 1C1 (AKR1C1) As the First Mutated Gene in a Family with Nonsyndromic Primary Lipedema
International Journal of Molecular Sciences Article Aldo-Keto Reductase 1C1 (AKR1C1) as the First Mutated Gene in a Family with Nonsyndromic Primary Lipedema Sandro Michelini 1 , Pietro Chiurazzi 2,3, Valerio Marino 4 , Daniele Dell’Orco 4 , Elena Manara 5 , Mirko Baglivo 5, Alessandro Fiorentino 1, Paolo Enrico Maltese 6 , Michele Pinelli 7,8 , Karen Louise Herbst 9, Astrit Dautaj 10 and Matteo Bertelli 5,10,* 1 Dipartimento di Riabilitazione, Ospedale San Giovanni Battista, A.C.I.S.M.O.M., 00148 Rome, Italy; [email protected] (S.M.); a.fi[email protected] (A.F.) 2 Istituto di Medicina Genomica, Università Cattolica del Sacro Cuore, 00168 Rome, Italy; [email protected] 3 Fondazione Policlinico Universitario “A.Gemelli” IRCCS, UOC Genetica Medica, 00168 Rome, Italy 4 Dipartimento di Neuroscienze, Biomedicina e Movimento, Sezione di Chimica Biologica, Università di Verona, 37134 Verona, Italy; [email protected] (V.M.); [email protected] (D.D.) 5 MAGI Euregio, 39100 Bolzano, Italy; [email protected] (E.M.); [email protected] (M.B.) 6 MAGI’s LAB, 38068 Rovereto, Italy; [email protected] 7 Dipartimento di Scienze Mediche Traslazionali, Sezione di Pediatria, Università di Napoli Federico II, 80131 Naples, Italy; [email protected] 8 Telethon Institute of Genetics and Medicine (TIGEM), 80078 Pozzuoli, Italy 9 Departments of Medicine, Pharmacy, Medical Imaging, Division of Endocrinology, University of Arizona, Tucson, AZ 85721, USA; [email protected] 10 EBTNA-Lab, 38068 Rovereto, Italy; [email protected] * Correspondence: [email protected] Received: 20 July 2020; Accepted: 27 August 2020; Published: 29 August 2020 Abstract: Lipedema is an often underdiagnosed chronic disorder that affects subcutaneous adipose tissue almost exclusively in women, which leads to disproportionate fat accumulation in the lower and upper body extremities. -

AKR1C1 As a Biomarker for Differentiating the Biological Effects of Combustible from Non-Combustible Tobacco Products
G C A T T A C G G C A T genes Article AKR1C1 as a Biomarker for Differentiating the Biological Effects of Combustible from Non-Combustible Tobacco Products Sangsoon Woo 1,†, Hong Gao 2,†, David Henderson 1, Wolfgang Zacharias 2,3, Gang Liu 4, Quynh T. Tran 4,* and G.L. Prasad 4 1 Statistical Genetics, Axio Research LLC, 4th Ave. Suite 200, Seattle, WA 98121, USA; [email protected] (S.W.); [email protected] (D.H.) 2 Department of Medicine, James Graham Brown Cancer Center, University of Louisville School of Medicine, Louisville, KY 40202, USA; [email protected] (H.G.); [email protected] (W.Z.) 3 Department of Pharmacology and Toxicology, University of Louisville School of Medicine, Louisville, KY 40202, USA 4 RAI Services Company, 401 N. Main Street, Winston-Salem, NC 27101, USA; [email protected] (G.L.); [email protected] (G.L.P.) * Correspondence: [email protected]; Tel.: +1-336-741-2772 † Co-first authors. Academic Editor: Selvarangan Ponnazhagan Received: 24 January 2017; Accepted: 25 April 2017; Published: 3 May 2017 Abstract: Smoking has been established as a major risk factor for developing oral squamous cell carcinoma (OSCC), but less attention has been paid to the effects of smokeless tobacco products. Our objective is to identify potential biomarkers to distinguish the biological effects of combustible tobacco products from those of non-combustible ones using oral cell lines. Normal human gingival epithelial cells (HGEC), non-metastatic (101A) and metastatic (101B) OSCC cell lines were exposed to different tobacco product preparations (TPPs) including cigarette smoke total particulate matter (TPM), whole-smoke conditioned media (WS-CM), smokeless tobacco extract in complete artificial saliva (STE), or nicotine (NIC) alone.