Anaerobe 2020 Congress Program & Abstract Book
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomics 98 (2011) 370–375
Genomics 98 (2011) 370–375 Contents lists available at ScienceDirect Genomics journal homepage: www.elsevier.com/locate/ygeno Whole-genome comparison clarifies close phylogenetic relationships between the phyla Dictyoglomi and Thermotogae Hiromi Nishida a,⁎, Teruhiko Beppu b, Kenji Ueda b a Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan b Life Science Research Center, College of Bioresource Sciences, Nihon University, Fujisawa, Japan article info abstract Article history: The anaerobic thermophilic bacterial genus Dictyoglomus is characterized by the ability to produce useful Received 2 June 2011 enzymes such as amylase, mannanase, and xylanase. Despite the significance, the phylogenetic position of Accepted 1 August 2011 Dictyoglomus has not yet been clarified, since it exhibits ambiguous phylogenetic positions in a single gene Available online 7 August 2011 sequence comparison-based analysis. The number of substitutions at the diverging point of Dictyoglomus is insufficient to show the relationships in a single gene comparison-based analysis. Hence, we studied its Keywords: evolutionary trait based on whole-genome comparison. Both gene content and orthologous protein sequence Whole-genome comparison Dictyoglomus comparisons indicated that Dictyoglomus is most closely related to the phylum Thermotogae and it forms a Bacterial systematics monophyletic group with Coprothermobacter proteolyticus (a constituent of the phylum Firmicutes) and Coprothermobacter proteolyticus Thermotogae. Our findings indicate that C. proteolyticus does not belong to the phylum Firmicutes and that the Thermotogae phylum Dictyoglomi is not closely related to either the phylum Firmicutes or Synergistetes but to the phylum Thermotogae. © 2011 Elsevier Inc. -

The Ecology of Staphylococcus Species in the Oral Cavity
J. Med. Microbiol. Ð Vol. 50 2001), 940±946 # 2001 The Pathological Society of Great Britain and Ireland ISSN 0022-2615 REVIEW ARTICLE The ecology of Staphylococcus species in the oral cavity A. J. SMITH, M. S. JACKSON and J. BAGG Infection Research Group, Glasgow Dental Hospital and School, 378 Sauchiehall Street, Glasgow G2 3JZ Whilst the diversity of organisms present in the oral cavity is well accepted, there remains considerable controversy as to whether Staphylococcus spp. play a role in the ecology of the normal oral ¯ora. Surprisingly little detailed work has been performed on the quantitative and qualitative aspects of colonisation or infection either by coagulase- negative staphylococci CNS) or S. aureus. The latter is especially interesting in the light of present dif®culties in eradicating carriage of methicillin-resistant S. aureus MRSA) from the oropharynx in affected individuals. This paper reviews the current knowledge of staphylococcal colonisation and infection of the oral cavity in health and disease. S. aureus has been isolated from a wide range of infective oral conditions, such as angular cheilitis and parotitis. More recently, a clinical condition classi®ed as staphylococcal mucositis has emerged as a clinical problem in many debilitated elderly patients and those with oral Crohn's disease. Higher carriage rates of both CNS or S. aureus,or both, in patients prone to joint infections raises the interesting possibility of the oral cavity serving as a potential source for bacteraemic spread to compromised joint spaces. In conclusion, there is a surprising paucity of knowledge regarding the role of oral staphylococci in both health and disease. -

And Methane-Oxidizing Microorganisms in a Dutch Drinking Water Treatment Plant
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.19.103440; this version posted May 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Metagenomic profiling of ammonia- and methane-oxidizing microorganisms in a Dutch drinking water treatment plant Lianna Poghosyan, Hanna Koch, Jeroen Frank, Maartje A.H.J. van Kessel, Geert Cremers, Theo van Alen, Mike S.M. Jetten, Huub J.M. Op den Camp, Sebastian Lücker* 5 Department of Microbiology, Radboud University, Heyendaalseweg 135, 6525 AJ Nijmegen, the Netherlands *Correspondence: [email protected] Keywords Sand filtration; nitrification; comammox Nitrospira; methanotrophic bacteria; metagenomics 10 Highlights • Microbial distribution was mainly influenced by sampling location within the DWTP. • Clade A comammox Nitrospira were the most abundant nitrifying guild in samples from the primary sand filter, while clade B dominated in samples from wall biofilm and the secondary filter. 15 • A novel methanotrophic bacterium affiliated with the Methylophilaceae family comprised the largest bacterial fraction in the primary sand filter. bioRxiv preprint doi: https://doi.org/10.1101/2020.05.19.103440; this version posted May 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Elevated concentrations of ammonium and methane in groundwater can cause severe problems during drinking water production. -

WO 2018/064165 A2 (.Pdf)
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/064165 A2 05 April 2018 (05.04.2018) W !P O PCT (51) International Patent Classification: Published: A61K 35/74 (20 15.0 1) C12N 1/21 (2006 .01) — without international search report and to be republished (21) International Application Number: upon receipt of that report (Rule 48.2(g)) PCT/US2017/053717 — with sequence listing part of description (Rule 5.2(a)) (22) International Filing Date: 27 September 2017 (27.09.2017) (25) Filing Language: English (26) Publication Langi English (30) Priority Data: 62/400,372 27 September 2016 (27.09.2016) US 62/508,885 19 May 2017 (19.05.2017) US 62/557,566 12 September 2017 (12.09.2017) US (71) Applicant: BOARD OF REGENTS, THE UNIVERSI¬ TY OF TEXAS SYSTEM [US/US]; 210 West 7th St., Austin, TX 78701 (US). (72) Inventors: WARGO, Jennifer; 1814 Bissonnet St., Hous ton, TX 77005 (US). GOPALAKRISHNAN, Vanch- eswaran; 7900 Cambridge, Apt. 10-lb, Houston, TX 77054 (US). (74) Agent: BYRD, Marshall, P.; Parker Highlander PLLC, 1120 S. Capital Of Texas Highway, Bldg. One, Suite 200, Austin, TX 78746 (US). (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Alteration of the Immune Response and the Microbiota of the Skin During a Natural Infection by Vibrio Harveyi in European Seabass (Dicentrarchus Labrax)
microorganisms Article Alteration of the Immune Response and the Microbiota of the Skin during a Natural Infection by Vibrio harveyi in European Seabass (Dicentrarchus labrax) María Cámara-Ruiz 1, Isabel M. Cerezo 2 , Francisco A. Guardiola 1 , José María García-Beltrán 1, M. Carmen Balebona 2 , Miguel Ángel Moriñigo 2 and María Ángeles Esteban 1,* 1 Immunobiology for Aquaculture Group, Department of Cell Biology and Histology, Faculty of Biology, Campus Regional de Excelencia Internacional “Campus Mare Nostrum”, University of Murcia, 30100 Murcia, Spain; [email protected] (M.C.-R.); [email protected] (F.A.G.); [email protected] (J.M.G.-B.) 2 Departamento de Microbiología, Universidad de Málaga, 29071 Málaga, Spain; [email protected] (I.M.C.); [email protected] (M.C.B.); [email protected] (M.Á.M.) * Correspondence: [email protected] Abstract: Disease outbreaks continue to represent one of the main bottlenecks for the sustainable development of the aquaculture industry. In marine aquaculture, many species from the Vibrio genus are serious opportunistic pathogens responsible for significant losses to producers. In this study, the effects on the immune response and the skin microbiota of European sea bass (Dicentrarchus labrax) were studied after a natural disease outbreak caused by V. harveyi. Data obtained from infected Citation: Cámara-Ruiz, M.; and non-infected fish were studied and compared. Regarding the local immune response (skin Cerezo, I.M.; Guardiola, F.A.; mucus) a decrease in the protease activity was observed in infected fish. Meanwhile, at a systemic García-Beltrán, J.M.; Balebona, M.C.; level, a decrease in protease and lysozyme activity was reported while peroxidase activity showed a Moriñigo, M.Á.; Esteban, M.Á. -

The C-Di-Gmp Regulatory Network in Clostridioides Difficile and Its Role in Modulating Surface Adherence and Persistence in the Mammalian Gut
THE C-DI-GMP REGULATORY NETWORK IN CLOSTRIDIOIDES DIFFICILE AND ITS ROLE IN MODULATING SURFACE ADHERENCE AND PERSISTENCE IN THE MAMMALIAN GUT Robert Woodrow McKee A dissertation submitted to the faculty at the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor in Philosophy in the Department of Microbiology and Immunology. Chapel Hill 2018 Approved by: Peggy A Cotter Jonathan J. Hansen Virginia L. Miller Rita Tamayo Mathew C Wolfgang © 2018 Robert Woodrow McKee ALL RIGHTS RESERVED ii ABSTRACT Robert Woodrow McKee: The c-di-GMP regulatory network in Clostridioides difficile and its role in modulating surface adherence and persistence in the mammalian gut (Under the direction of Rita Tamayo) Clostridioides difficile (Clostridium difficile) is a spore-forming bacterial pathogen responsible for hundreds of thousands of infections each year in the United States. C. difficile outbreaks are common in hospitals because C. difficile spores can persist for months on surfaces and are resistant to many disinfectants. Despite the significant disease burden that C. difficile represents, we know surprisingly little about the factors necessary for C. difficile to colonize and persist in the mammalian intestine. Previous work demonstrated that the signaling molecule cyclic diguanylate (c-di-GMP) regulates a variety of processes in C. difficile including production of the toxins that are required for disease symptoms. Using monolayers of human intestinal epithelial cells, we demonstrate that c-di-GMP promotes attachment of C. difficile to intestinal epithelial cells. We also demonstrate that regulation of type IV pili (TFP) by c-di-GMP promotes prolonged adherence of C. -

Table S4. Phylogenetic Distribution of Bacterial and Archaea Genomes in Groups A, B, C, D, and X
Table S4. Phylogenetic distribution of bacterial and archaea genomes in groups A, B, C, D, and X. Group A a: Total number of genomes in the taxon b: Number of group A genomes in the taxon c: Percentage of group A genomes in the taxon a b c cellular organisms 5007 2974 59.4 |__ Bacteria 4769 2935 61.5 | |__ Proteobacteria 1854 1570 84.7 | | |__ Gammaproteobacteria 711 631 88.7 | | | |__ Enterobacterales 112 97 86.6 | | | | |__ Enterobacteriaceae 41 32 78.0 | | | | | |__ unclassified Enterobacteriaceae 13 7 53.8 | | | | |__ Erwiniaceae 30 28 93.3 | | | | | |__ Erwinia 10 10 100.0 | | | | | |__ Buchnera 8 8 100.0 | | | | | | |__ Buchnera aphidicola 8 8 100.0 | | | | | |__ Pantoea 8 8 100.0 | | | | |__ Yersiniaceae 14 14 100.0 | | | | | |__ Serratia 8 8 100.0 | | | | |__ Morganellaceae 13 10 76.9 | | | | |__ Pectobacteriaceae 8 8 100.0 | | | |__ Alteromonadales 94 94 100.0 | | | | |__ Alteromonadaceae 34 34 100.0 | | | | | |__ Marinobacter 12 12 100.0 | | | | |__ Shewanellaceae 17 17 100.0 | | | | | |__ Shewanella 17 17 100.0 | | | | |__ Pseudoalteromonadaceae 16 16 100.0 | | | | | |__ Pseudoalteromonas 15 15 100.0 | | | | |__ Idiomarinaceae 9 9 100.0 | | | | | |__ Idiomarina 9 9 100.0 | | | | |__ Colwelliaceae 6 6 100.0 | | | |__ Pseudomonadales 81 81 100.0 | | | | |__ Moraxellaceae 41 41 100.0 | | | | | |__ Acinetobacter 25 25 100.0 | | | | | |__ Psychrobacter 8 8 100.0 | | | | | |__ Moraxella 6 6 100.0 | | | | |__ Pseudomonadaceae 40 40 100.0 | | | | | |__ Pseudomonas 38 38 100.0 | | | |__ Oceanospirillales 73 72 98.6 | | | | |__ Oceanospirillaceae -

Extensive Microbial Diversity Within the Chicken Gut Microbiome Revealed by Metagenomics and Culture
Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture Rachel Gilroy1, Anuradha Ravi1, Maria Getino2, Isabella Pursley2, Daniel L. Horton2, Nabil-Fareed Alikhan1, Dave Baker1, Karim Gharbi3, Neil Hall3,4, Mick Watson5, Evelien M. Adriaenssens1, Ebenezer Foster-Nyarko1, Sheikh Jarju6, Arss Secka7, Martin Antonio6, Aharon Oren8, Roy R. Chaudhuri9, Roberto La Ragione2, Falk Hildebrand1,3 and Mark J. Pallen1,2,4 1 Quadram Institute Bioscience, Norwich, UK 2 School of Veterinary Medicine, University of Surrey, Guildford, UK 3 Earlham Institute, Norwich Research Park, Norwich, UK 4 University of East Anglia, Norwich, UK 5 Roslin Institute, University of Edinburgh, Edinburgh, UK 6 Medical Research Council Unit The Gambia at the London School of Hygiene and Tropical Medicine, Atlantic Boulevard, Banjul, The Gambia 7 West Africa Livestock Innovation Centre, Banjul, The Gambia 8 Department of Plant and Environmental Sciences, The Alexander Silberman Institute of Life Sciences, Edmond J. Safra Campus, Hebrew University of Jerusalem, Jerusalem, Israel 9 Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield, UK ABSTRACT Background: The chicken is the most abundant food animal in the world. However, despite its importance, the chicken gut microbiome remains largely undefined. Here, we exploit culture-independent and culture-dependent approaches to reveal extensive taxonomic diversity within this complex microbial community. Results: We performed metagenomic sequencing of fifty chicken faecal samples from Submitted 4 December 2020 two breeds and analysed these, alongside all (n = 582) relevant publicly available Accepted 22 January 2021 chicken metagenomes, to cluster over 20 million non-redundant genes and to Published 6 April 2021 construct over 5,500 metagenome-assembled bacterial genomes. -

Pathogen Challenge and Dietary Shift Alter Microbiota Composition And
fmicb-12-703421 July 19, 2021 Time: 11:40 # 1 ORIGINAL RESEARCH published: 19 July 2021 doi: 10.3389/fmicb.2021.703421 Pathogen Challenge and Dietary Shift Alter Microbiota Composition and Activity in a Mucin-Associated in vitro Model of the Piglet Colon (MPigut-IVM) Simulating Weaning Transition Raphaële Gresse1,2, Frédérique Chaucheyras-Durand1,2, Juan J. Garrido3, Sylvain Denis1, Angeles Jiménez-Marín3, Martin Beaumont4, Tom Van de Wiele5, Evelyne Forano1 and Stéphanie Blanquet-Diot1* 1 INRAE, UMR 454 MEDIS, Université Clermont Auvergne, Clermont-Ferrand, France, 2 Lallemand SAS, Blagnac, France, 3 Grupo de Genómica y Mejora Animal, Departamento de Genética, Facultad de Veterinaria, Universidad de Córdoba, Córdoba, Spain, 4 GenPhySE, INRAE, ENVT, Université de Toulouse, Castanet-Tolosan, France, 5 Center for Microbial Ecology and Technology, Ghent University, Ghent, Belgium Edited by: Wakako Ikeda-Ohtsubo, Tohoku University, Japan Enterotoxigenic Escherichia coli (ETEC) is the principal pathogen responsible for post- Reviewed by: weaning diarrhea in newly weaned piglets. Expansion of ETEC at weaning is thought to Katie Lynn Summers, be the consequence of various stress factors such as transient anorexia, dietary change United States Department or increase in intestinal inflammation and permeability, but the exact mechanisms remain of Agriculture (USDA), United States Åsa Sjöling, to be elucidated. As the use of animal experiments raise more and more ethical Karolinska Institutet (KI), Sweden concerns, we used a recently developed in vitro model of piglet colonic microbiome *Correspondence: and mucobiome, the MPigut-IVM, to evaluate the effects of a simulated weaning Stéphanie Blanquet-Diot [email protected] transition and pathogen challenge at weaning. -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

Common Commensals
Common Commensals Actinobacterium meyeri Aerococcus urinaeequi Arthrobacter nicotinovorans Actinomyces Aerococcus urinaehominis Arthrobacter nitroguajacolicus Actinomyces bernardiae Aerococcus viridans Arthrobacter oryzae Actinomyces bovis Alpha‐hemolytic Streptococcus, not S pneumoniae Arthrobacter oxydans Actinomyces cardiffensis Arachnia propionica Arthrobacter pascens Actinomyces dentalis Arcanobacterium Arthrobacter polychromogenes Actinomyces dentocariosus Arcanobacterium bernardiae Arthrobacter protophormiae Actinomyces DO8 Arcanobacterium haemolyticum Arthrobacter psychrolactophilus Actinomyces europaeus Arcanobacterium pluranimalium Arthrobacter psychrophenolicus Actinomyces funkei Arcanobacterium pyogenes Arthrobacter ramosus Actinomyces georgiae Arthrobacter Arthrobacter rhombi Actinomyces gerencseriae Arthrobacter agilis Arthrobacter roseus Actinomyces gerenseriae Arthrobacter albus Arthrobacter russicus Actinomyces graevenitzii Arthrobacter arilaitensis Arthrobacter scleromae Actinomyces hongkongensis Arthrobacter astrocyaneus Arthrobacter sulfonivorans Actinomyces israelii Arthrobacter atrocyaneus Arthrobacter sulfureus Actinomyces israelii serotype II Arthrobacter aurescens Arthrobacter uratoxydans Actinomyces meyeri Arthrobacter bergerei Arthrobacter ureafaciens Actinomyces naeslundii Arthrobacter chlorophenolicus Arthrobacter variabilis Actinomyces nasicola Arthrobacter citreus Arthrobacter viscosus Actinomyces neuii Arthrobacter creatinolyticus Arthrobacter woluwensis Actinomyces odontolyticus Arthrobacter crystallopoietes -
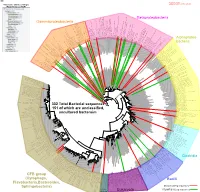
Bacteria Clostridia Bacilli Eukaryota CFB Group
AM935842.1.1361 uncultured Burkholderiales bacterium Class Betaproteobacteria AY283260.1.1552 Alcaligenes sp. PCNB−2 Class Betaproteobacteria AM934953.1.1374 uncultured Burkholderiales bacterium Class Betaproteobacteria AJ581593.1.1460 uncultured betaAM936569.1.1351 proteobacterium uncultured Class Betaproteobacteria Derxia sp. Class Betaproteobacteria AJ581621.1.1418 uncultured beta proteobacterium Class Betaproteobacteria DQ248272.1.1498 uncultured soil bacterium soil uncultured DQ248272.1.1498 DQ248235.1.1498 uncultured soil bacterium RS49 DQ248270.1.1496 uncultured soil bacterium DQ256489.1.1211 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax DQ256489.1.1211 AF523053.1.1486 uncultured Comamonadaceae bacterium Class Betaproteobacteria AY706442.1.1396 uncultured bacterium uncultured AY706442.1.1396 AJ536763.1.1422 uncultured bacterium CS000359.1.1530 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax CS000359.1.1530 AY168733.1.1411 uncultured bacterium AJ009470.1.1526 uncultured bacterium SJA−62 Class Betaproteobacteria Class SJA−62 bacterium uncultured AJ009470.1.1526 AY212561.1.1433 uncultured bacterium D16212.1.1457 Rhodoferax fermentans Class Betaproteobacteria Class fermentans Rhodoferax D16212.1.1457 AY957894.1.1546 uncultured bacterium AJ581620.1.1452 uncultured beta proteobacterium Class Betaproteobacteria RS76 AY625146.1.1498 uncultured bacterium RS65 DQ316832.1.1269 uncultured beta proteobacterium Class Betaproteobacteria DQ404909.1.1513 uncultured bacterium uncultured DQ404909.1.1513 AB021341.1.1466 bacterium rM6 AJ487020.1.1500 uncultured bacterium uncultured AJ487020.1.1500 RS7 RS86RC AF364862.1.1425 bacterium BA128 Class Betaproteobacteria AY957931.1.1529 uncultured bacterium uncultured AY957931.1.1529 CP000884.723807.725332 Delftia acidovorans SPH−1 Class Betaproteobacteria AY957923.1.1520 uncultured bacterium uncultured AY957923.1.1520 RS18 AY957918.1.1527 uncultured bacterium uncultured AY957918.1.1527 AY945883.1.1500 uncultured bacterium AF526940.1.1489 uncultured Ralstonia sp.