Detection of Bacterial Retroelements Using Genomics Sen Mu East Tennessee State University
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Two Independent Retrons with Highly Diverse Reverse Transcriptases In
Proc. Natl. Acad. Sci. USA Vol. 87, pp. 942-945, February 1990 Biochemistry Two independent retrons with highly diverse reverse transcriptases in Myxococcus xanthus (multicopy single-stranded DNA/2',5'-phosphodiester/codon usage/myxobacteria) SUMIKO INOUYE, PETER J. HERZER, AND MASAYORI INOUYE Department of Biochemistry, University of Medicine and Dentistry of New Jersey, Robert Wood Johnson Medical School, Piscataway, NJ 08854 Communicated by Russell F. Doolittle, November 27, 1989 (received for review September 29, 1989) ABSTRACT A reverse transcriptase (RT) was recently found that the Mx65 retron is highly diverse and independent found in Myxococcus xanthus, a Gram-negative soil bacterium. from the Mx162 retron and that RT associated with the Mx65 This RT has been shown to be associated with a chromosomal retron has only 47% identity with RT for the Mx162 retron. region designated a retron responsible for the synthesis of a peculiar extrachromosomal DNA called msDNA (multicopy single-stranded DNA). We demonstrate that M. xanthus con- MATERIALS AND METHODS tains two independent, unlinked retrons, one for the synthesis Materials. The clone of the 9.0-kilobase (kb) Pst I fragment of msDNA-Mxl62 and the other for msDNA-Mx65. The struc- containing the Mx65 retron was obtained (8). Restriction tural analysis of the retron for msDNA-Mx65 revealed that the enzymes were purchased from New England Biolabs and coding regions for msdRNA (msr) and msDNA (msd), and an Boehringer Mannheim. open reading frame (ORF) downstream of msr are arranged in A deletion mutant strain ofthe Mx162 retron, AmsSX, was the same manner as found for the Mx162 retron. -
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
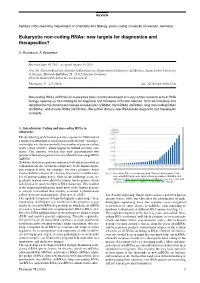
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

First Genomic Insights Into Members of a Candidate Bacterial Phylum Responsible for Wastewater Bulking
First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking Yuji Sekiguchi1, Akiko Ohashi1, Donovan H. Parks2, Toshihiro Yamauchi3, Gene W. Tyson2,4 and Philip Hugenholtz2,5 1 Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Ibaraki, Japan 2 Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, The University of Queensland, St. Lucia, Queensland, Australia 3 Administrative Management Department, Kubota Kasui Corporation, Minato-ku, Tokyo, Japan 4 Advanced Water Management Centre, The University of Queensland, St. Lucia, Queensland, Australia 5 Institute for Molecular Bioscience, The University of Queensland, St. Lucia, Queensland, Australia ABSTRACT Filamentous cells belonging to the candidate bacterial phylum KSB3 were previously identified as the causative agent of fatal filament overgrowth (bulking) in a high-rate industrial anaerobic wastewater treatment bioreactor. Here, we obtained near complete genomes from two KSB3 populations in the bioreactor, including the dominant bulking filament, using diVerential coverage binning of metagenomic data. Fluorescence in situ hybridization with 16S rRNA-targeted probes specific for the two populations confirmed that both are filamentous organisms. Genome-based metabolic reconstruction and microscopic observation of the KSB3 filaments in the presence of sugar gradients indicate that both filament types are Gram-negative, strictly anaerobic fermenters capable of -

A Pirna-Like Small RNA Interacts with and Modulates P-ERM Proteins in Human Somatic Cells
ARTICLE Received 18 Mar 2015 | Accepted 27 Apr 2015 | Published 22 June 2015 DOI: 10.1038/ncomms8316 OPEN A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells Yuping Mei1, Yuyan Wang1,2, Priti Kumari3, Amol Carl Shetty3, David Clark1, Tyler Gable1, Alexander D. MacKerell4,5, Mark Z. Ma1, David J. Weber5,6, Austin J. Yang5, Martin J. Edelman5 & Li Mao1,5 PIWI-interacting RNAs (piRNAs) are thought to silence transposon and gene expression during development. However, the roles of piRNAs in somatic tissues are largely unknown. Here we report the identification of 555 piRNAs in human lung bronchial epithelial (HBE) and non-small cell lung cancer (NSCLC) cell lines, including 295 that do not exist in databases termed as piRNA-like sncRNAs or piRNA-Ls. Distinctive piRNA/piRNA-L expression patterns are observed between HBE and NSCLC cells. piRNA-like-163 (piR-L-163), the top down- regulated piRNA-L in NSCLC cells, binds directly to phosphorylated ERM proteins (p-ERM), which is dependent on the central part of UUNNUUUNNUU motif in piR-L-163 and the RRRKPDT element in ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions. 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, 650W Baltimore Street, Baltimore, Maryland 21201, USA. 2 Department of Thoracic Medical Oncology, Peking University Cancer Hospital and Institute, Beijing 100142, China. -

RNA-Based Technologies for Engineering Plant Virus Resistance
plants Review RNA-Based Technologies for Engineering Plant Virus Resistance Michael Taliansky 1,2,*, Viktoria Samarskaya 1, Sergey K. Zavriev 1 , Igor Fesenko 1 , Natalia O. Kalinina 1,3 and Andrew J. Love 2,* 1 Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, 117997 Moscow, Russia; [email protected] (V.S.); [email protected] (S.K.Z.); [email protected] (I.F.); [email protected] (N.O.K.) 2 The James Hutton Institute, Invergowrie, Dundee DD2 5DA, UK 3 Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Leninskie Gory, 119991 Moscow, Russia * Correspondence: [email protected] (M.T.); [email protected] (A.J.L.) Abstract: In recent years, non-coding RNAs (ncRNAs) have gained unprecedented attention as new and crucial players in the regulation of numerous cellular processes and disease responses. In this review, we describe how diverse ncRNAs, including both small RNAs and long ncRNAs, may be used to engineer resistance against plant viruses. We discuss how double-stranded RNAs and small RNAs, such as artificial microRNAs and trans-acting small interfering RNAs, either produced in transgenic plants or delivered exogenously to non-transgenic plants, may constitute powerful RNA interference (RNAi)-based technology that can be exploited to control plant viruses. Additionally, we describe how RNA guided CRISPR-CAS gene-editing systems have been deployed to inhibit plant virus infections, and we provide a comparative analysis of RNAi approaches and CRISPR-Cas technology. The two main strategies for engineering virus resistance are also discussed, including direct targeting of viral DNA or RNA, or inactivation of plant host susceptibility genes. -

The Myxocoumarins a and B from Stigmatella Aurantiaca Strain MYX-030
The myxocoumarins A and B from Stigmatella aurantiaca strain MYX-030 Tobias A. M. Gulder*1, Snežana Neff2, Traugott Schüz2, Tammo Winkler2, René Gees2 and Bettina Böhlendorf*2 Full Research Paper Open Access Address: Beilstein J. Org. Chem. 2013, 9, 2579–2585. 1Kekulé Institute of Organic Chemistry and Biochemistry, University of doi:10.3762/bjoc.9.293 Bonn, Gerhard-Domagk-Straβe 1, 53121 Bonn, Germany and 2Syngenta Crop Protection AG, CH-4002 Basel, Switzerland Received: 21 August 2013 Accepted: 01 November 2013 Email: Published: 20 November 2013 Tobias A. M. Gulder* - [email protected]; Bettina Böhlendorf* - [email protected] This article is part of the Thematic Series "Natural products in synthesis and biosynthesis" and is dedicated to Prof. Dr. Gerhard Höfle on the * Corresponding author occasion of his 74th birthday. Keywords: Guest Editor: J. S. Dickschat antifungal activity; myxobacteria; natural products; Stigmatella aurantiaca; structure elucidation © 2013 Gulder et al; licensee Beilstein-Institut. License and terms: see end of document. Abstract The myxobacterial strain Stigmatella aurantiaca MYX-030 was selected as promising source for the discovery of new biologically active natural products by our screening methodology. The isolation, structure elucidation and initial biological evaluation of the myxocoumarins derived from this strain are described in this work. These compounds comprise an unusual structural framework and exhibit remarkable antifungal properties. Introduction Despite declining interest of most big R&D-driven chemical epothilones A (1), Figure 1), microtubule-stabilizing macrolac- companies in recent years, natural products continue to serve as tones that are clinically used in cancer therapy [16-20]. one of the most important sources of new bioactive chemical entities, both in the pharmaceutical [1-4] as well as in the agro- But also from an agrochemical point of view, myxobacteria chemical industry [5-8]. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Questions with Answers- Nucleotides & Nucleic Acids A. the Components
Questions with Answers- Nucleotides & Nucleic Acids A. The components and structures of common nucleotides are compared. (Questions 1-5) 1._____ Which structural feature is shared by both uracil and thymine? a) Both contain two keto groups. b) Both contain one methyl group. c) Both contain a five-membered ring. d) Both contain three nitrogen atoms. 2._____ Which component is found in both adenosine and deoxycytidine? a) Both contain a pyranose. b) Both contain a 1,1’-N-glycosidic bond. c) Both contain a pyrimidine. d) Both contain a 3’-OH group. 3._____ Which property is shared by both GDP and AMP? a) Both contain the same charge at neutral pH. b) Both contain the same number of phosphate groups. c) Both contain the same purine. d) Both contain the same furanose. 4._____ Which characteristic is shared by purines and pyrimidines? a) Both contain two heterocyclic rings with aromatic character. b) Both can form multiple non-covalent hydrogen bonds. c) Both exist in planar configurations with a hemiacetal linkage. d) Both exist as neutral zwitterions under cellular conditions. 5._____ Which property is found in nucleosides and nucleotides? a) Both contain a nitrogenous base, a pentose, and at least one phosphate group. b) Both contain a covalent phosphodister bond that is broken in strong acid. c) Both contain an anomeric carbon atom that is part of a β-N-glycosidic bond. d) Both contain an aldose with hydroxyl groups that can tautomerize. ___________________________________________________________________________ B. The structures of nucleotides and their components are studied. (Questions 6-10) 6._____ Which characteristic is shared by both adenine and cytosine? a) Both contain one methyl group. -

Inactivation of Stigmatella Aurantiaca Csga Gene Impares Rippling Formation- Genetika, Vol 47, No
UDC 575 DOI: 10.2298/GENSR1501119M Original scientific paper INACTIVATION OF Stigmatella aurantiaca csg A GENE IMPARES RIPPLING FORMATION Ana MILOSEVIC-DJERIC 1, Susanne MÜLLER 2, and Hans URLICH SCHAIRER 3 1 Department of Gynaecology and Obstetrics, Hospital Centre Uzice, Uzice, Serbia 2 Univeristy of Iowa, Iowa City, USA 3Zentrum fur Molekulare Biologie, ZMBH, Heidelberg, Germany Milosevic-Djeric A., S. Müller, and H. Urlich Schairer (2015): Inactivation of Stigmatella aurantiaca csgA gene impares rippling formation- Genetika, Vol 47, No. 1, 119-130. Stigmatella aurantiaca fruiting body development depends on cell-cell interactions. One type of the signaling molecule stigmolone isolated from S. aurantiaca cells acts to help cells to stay together in the aggregation phase. Another gene product involved in intercellular signaling in S. aurantiaca is the csg A homolog of Myxococcus xanthus . In close relative M. xanthus C signal the product of the csg A gene is required for rippling, aggregation and sporulation. Isolation of homologous gene in S. aurantiaca implicates a probable role of CsgA in intercellular communication. Inactivation of the gene by insertion mutagenesis caused alterations in S. aurantiaca fruiting. The motility behavior of the cells during development was changed as well as their ability to stay more closely together in the early stages of development. Inactivation of the csg A gene completely abolished rippling of the cells. This indicates the crucial role of the CsgA protein in regulating this rhythmic behavior. Key words: cell-cell signaling, development, fruiting body formation, myxobacteria, Stigmatella aurantiaca INTRODUCTION Stigmatella aurantiaca belongs to the myxobacteria famous for their ability to form multicellular structures fruiting bodies in response to the starvation conditions ( SHIMKETS et al ., 2006; ZHANG et al., 2012; CLAESSEN et al ., 2014). -

Mirna Detection Using a Rolling Circle Amplification and RNA
biosensors Article MiRNA Detection Using a Rolling Circle Amplification and RNA-Cutting Allosteric Deoxyribozyme Dual Signal Amplification Strategy Chenxin Fang, Ping Ouyang, Yuxing Yang, Yang Qing, Jialun Han, Wenyan Shang, Yubing Chen and Jie Du * State Key Laboratory of Marine Resource Utilization in South China Sea, College of Materials Science and Engineering, Hainan University, Haikou 570228, China; [email protected] (C.F.); [email protected] (P.O.); [email protected] (Y.Y.); [email protected] (Y.Q.); [email protected] (J.H.); [email protected] (W.S.); [email protected] (Y.C.) * Correspondence: [email protected] Abstract: A microRNA (miRNA) detection platform composed of a rolling circle amplification (RCA) system and an allosteric deoxyribozyme system is proposed, which can detect miRNA-21 rapidly and efficiently. Padlock probe hybridization with the target miRNA is achieved through complementary base pairing and the padlock probe forms a closed circular template under the action of ligase; this circular template results in RCA. In the presence of DNA polymerase, RCA proceeds and a long chain with numerous repeating units is formed. In the presence of single-stranded DNA (H1 and H2), multi-component nucleic acid enzymes (MNAzymes) are formed that have the ability to cleave substrates. Finally, substrates containing fluorescent and quenching groups and magnesium ions are added to the system to activate the MNAzyme and the substrate cleavage reaction, thus achieving fluorescence intensity amplification. The RCA–MNAzyme system has dual signal amplification and presents a sensing platform that demonstrates broad prospects in the analysis and detection of Citation: Fang, C.; Ouyang, P.; Yang, nucleic acids. -

Neo-Cloo2 B2 O 1 B 1 (3) REVERSE TRANSCRIPTION RNA PROCESSING
|||||||||||||| O US005436141A United States Patent (19) 11 Patent Number: 5,436,141 Miyata et al. (45) Date of Patent: Jul. 25, 1995 54 METHOD FOR SYNTHESIZING STABLE 56) References Cited SINGLE-STRANDED CONAN EUKARYOTES BY MEANS OF A U.S. PATENT DOCUMENTS BACTERAL RETRON AND PRODUCTS 5,079,151 1/1992 Lampson et al. ................ 435/91.51 75 Inventors: Shohei Miyata, Misato, Japan; FOREIGN PATENT DOCUMENTS Atsushi Ohshima, Highland Park, 0132309 1/1985 European Pat. Off. ... C12N 15/00 N.J.; Sumiko Inouye; Masayori Inouye, both of Bridgewater, N.J. OTHER PUBLICATIONS Y o Dhundale et al., J. Bact, vol. 170, 1988, pp. 5620-5624. 73) Assignee: University of Medicine and Dentistry Lampson et al., Science, vol. 243, 1989, pp. 1033-1038. of New Jersey, Newark, N.J. Sambrook et al., Molecular Cloning: A Laboratory Man ual, Cold Spring Harbor Laboratory Press, 1989, pp. 21 Appl. No.: 753,110 16.15-16.16. 22 Filed: Aug. 30, 1991 Primary Examiner-Richard A. Schwartz Assistant Examiner-James Ketter Related U.S. Application Data Attorney, Agent, or Firm-Weiser & Associates (63) Continuation-in-part of Ser. No. 315,427, Feb. 24, 1989, 57 ABSTRACT Pat. No. 5,979,151, and a continuation in part of Ser. A method for producing in vivo stable single-stranded No. 315,316, Feb. 24, 1989, Pat. No. 5,320,958, and a DNAs in eucaryotic cells. The DNAs are multicopy Eup ii.g 1. single-stranded DNA (msDNA) structures constituted 517,946, May 2, 1990, and a continuation-in-part of Ser. by a RNA and a DNA portion. -

Reverse Transcriptase Genes Are Highly Abundant and Transcriptionally Active in Marine Plankton Assemblages
The ISME Journal (2016) 10, 1134–1146 © 2016 International Society for Microbial Ecology All rights reserved 1751-7362/16 OPEN www.nature.com/ismej ORIGINAL ARTICLE Reverse transcriptase genes are highly abundant and transcriptionally active in marine plankton assemblages Magali Lescot1, Pascal Hingamp1, Kenji K Kojima2, Emilie Villar1, Sarah Romac3,4, Alaguraj Veluchamy5,11, Martine Boccara5, Olivier Jaillon6,7,8, Daniele Iudicone9, Chris Bowler5, Patrick Wincker6,7,8, Jean-Michel Claverie1 and Hiroyuki Ogata10 1Information Génomique et Structurale, UMR7256, CNRS, Aix-Marseille Université, Institut de Microbiologie de la Méditerranée (FR3479), Parc Scientifique de Luminy, Marseille, France; 2Genetic Information Research Institute, Los Altos, CA, USA; 3CNRS, UMR 7144, team EPEP, Station Biologique de Roscoff, Place Georges Teissier, Roscoff, France; 4Sorbonne Universités, UPMC Univ Paris 06, Station Biologique de Roscoff, Place Georges Teissier, FR-Roscoff, France; 5Ecole Normale Supérieure, PSL Research University, Institut de Biologie de l’Ecole Normale Supérieure (IBENS), Paris, France; 6CEA-Institut de Génomique, GENOSCOPE, Centre National de Séquençage, Evry Cedex, France; 7Université d’Evry, Evry Cedex, France; 8Centre National de la Recherche Scientifique (CNRS), Evry Cedex, France; 9Laboratory of Ecology and Evolution of Plankton, Stazione Zoologica Anton Dohrn, Naples, Italy and 10Bioinformatics Center, Institute for Chemical Research, Kyoto University, Gokasho, Uji, Kyoto, Japan Genes encoding reverse transcriptases (RTs) are found in most eukaryotes, often as a component of retrotransposons, as well as in retroviruses and in prokaryotic retroelements. We investigated the abundance, classification and transcriptional status of RTs based on Tara Oceans marine metagenomes and metatranscriptomes encompassing a wide organism size range. Our analyses revealed that RTs predominate large-size fraction metagenomes (45 μm), where they reached a maximum of 13.5% of the total gene abundance.