B a C D E F G
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Recombinant MAP2K2 Protein
Recombinant MAP2K2 protein Catalog No: 81332, 81632 Quantity: 20, 1000 µg Expressed In: Baculovirus Concentration: 0.3 µg/µl Source: Human Buffer Contents: Recombinant MAP2K2 protein is supplied in 25 mM HEPES-NaOH pH 7.5, 300 mM NaCl, 10% glycerol, 0.04% Triton X-100, and 0.5 mM TCEP. Background: MAP2K2 (Mitogen-Activated Protein Kinase Kinase Kinase 2) is a dual specificity protein kinase that belongs to the MAP kinase kinase family. This kinase plays a critical role in mitogen growth factor signal transduction. It phosphorylates and activates MAPK1/ERK2 and MAPK2/ERK3. The activation of this kinase itself is dependent on the Ser/Thr phosphorylation by MAP kinase kinase kinases. Mutations in this gene cause cardiofaciocutaneous syndrome (CFC syndrome), a disease characterized by heart defects, cognitive disability, and distinctive facial features similar to those found in Noonan syndrome. Protein Details: Recombinant MAP2K2 protein was expressed in baculovirus expression system as the full length protein (accession number NP_109587.1) with a N-terminal FLAG Tag. The molecular weight of the protein is 45.7 kDa. Application Notes: This product was manufactured as described in Protein Details. Where possible, Active Motif has developed functional or activity assays for Recombinant MAP2K2 protein gel recombinant proteins. Additional characterization such as enzyme kinetic activity 10% SDS-PAGE with Coomassie blue assays, inhibitor screening or other biological activity assays may not have been staining MW: 45.7 kDa performed for every product. All available data for a given product is shown on the lot- Purity: >90% specific Technical Data Sheet. Storage and Guarantee: Recombinant proteins in solution are temperature sensitive and must be stored at -80°C to prevent degradation. -

Clustering of the Structures of Protein Kinase Activation Loops
bioRxiv preprint doi: https://doi.org/10.1101/395723; this version posted August 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Clustering of the structures of protein kinase activation loops: A new nomenclature for active and inactive kinase structures Vivek Modi Roland L. Dunbrack, Jr.* Institute for Cancer Research Fox Chase Cancer Center Philadelphia PA 19111 *[email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/395723; this version posted August 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract TarGeting protein kinases is an important strateGy for intervention in cancer. Inhibitors are directed at the conserved active conformation or a variety of inactive conformations. While attempts have been made to classify these conformations, a structurally rigorous cataloGue of states has not been achieved. The kinase activation loop is crucial for catalysis and beGins with the conserved DFGmotif (Asp-Phe-Gly). This motif is observed in two major classes of conformations, DFGin - an ensemble of active and inactive conformations where the Phe residue is in contact with the C-helix of the N-terminal lobe, and DFGout - an inactive form where Phe occupies the ATP site exposinG the C-helix pocket. -

Targeting Fibrosis in the Duchenne Muscular Dystrophy Mice Model: an Uphill Battle
bioRxiv preprint doi: https://doi.org/10.1101/2021.01.20.427485; this version posted January 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Title: Targeting fibrosis in the Duchenne Muscular Dystrophy mice model: an uphill battle 2 Marine Theret1#, Marcela Low1#, Lucas Rempel1, Fang Fang Li1, Lin Wei Tung1, Osvaldo 3 Contreras3,4, Chih-Kai Chang1, Andrew Wu1, Hesham Soliman1,2, Fabio M.V. Rossi1 4 1School of Biomedical Engineering and the Biomedical Research Centre, Department of Medical 5 Genetics, 2222 Health Sciences Mall, Vancouver, BC, V6T 1Z3, Canada 6 2Department of Pharmacology and Toxicology, Faculty of Pharmaceutical Sciences, Minia 7 University, Minia, Egypt 8 3Developmental and Stem Cell Biology Division, Victor Chang Cardiac Research Institute, 9 Darlinghurst, NSW, 2010, Australia 10 4Departamento de Biología Celular y Molecular and Center for Aging and Regeneration (CARE- 11 ChileUC), Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, 8331150 12 Santiago, Chile 13 # Denotes Co-first authorship 14 15 Keywords: drug screening, fibro/adipogenic progenitors, fibrosis, repair, skeletal muscle. 16 Correspondence to: 17 Marine Theret 18 School of Biomedical Engineering and the Biomedical Research Centre 19 University of British Columbia 20 2222 Health Sciences Mall, Vancouver, British Columbia 21 Tel: +1(604) 822 0441 fax: +1(604) 822 7815 22 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.01.20.427485; this version posted January 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Map2k1 and Map2k2 Genes Contribute to the Normal Development of Syncytiotrophoblasts During Placentation
RESEARCH ARTICLE 1363 Development 136, 1363-1374 (2009) doi:10.1242/dev.031872 Map2k1 and Map2k2 genes contribute to the normal development of syncytiotrophoblasts during placentation Valérie Nadeau*, Stéphanie Guillemette*, Louis-François Bélanger, Olivier Jacob, Sophie Roy and Jean Charron† The mammalian genome contains two ERK/MAP kinase kinase genes, Map2k1 and Map2k2, which encode dual-specificity kinases responsible for ERK/MAP kinase activation. In the mouse, loss of Map2k1 function causes embryonic lethality, whereas Map2k2 mutants survive with a normal lifespan, suggesting that Map2k1 masks the phenotype due to the Map2k2 mutation. To uncover the specific function of MAP2K2 and the threshold requirement of MAP2K proteins during embryo formation, we have successively ablated the Map2k gene functions. We report here that Map2k2 haploinsufficiency affects the normal development of placenta in the absence of one Map2k1 allele. Most Map2k1+/–Map2k2+/– embryos die during gestation because of placenta defects restricted to extra-embryonic tissues. The impaired viability of Map2k1+/–Map2k2+/– embryos can be rescued when the Map2k1 deletion is restricted to the embryonic tissues. The severity of the placenta phenotype is dependent on the number of Map2k mutant alleles, the deletion of the Map2k1 allele being more deleterious. Moreover, the deletion of one or both Map2k2 alleles in the context of one null Map2k1 allele leads to the formation of multinucleated trophoblast giant (MTG) cells. Genetic experiments indicate that these structures are derived from Gcm1-expressing syncytiotrophoblasts (SynT), which are affected in their ability to form the uniform SynT layer II lining the maternal sinuses. Thus, even though Map2k1 plays a predominant role, these results enlighten the function of Map2k2 in placenta development. -

Human Kinome Profiling Identifies a Requirement for AMP-Activated
Human kinome profiling identifies a requirement for AMP-activated protein kinase during human cytomegalovirus infection Laura J. Terrya, Livia Vastagb,1, Joshua D. Rabinowitzb, and Thomas Shenka,2 aDepartment of Molecular Biology and bDepartment of Chemistry and the Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ 08544 Contributed by Thomas Shenk, January 11, 2012 (sent for review December 29, 2011) Human cytomegalovirus (HCMV) modulates numerous cellular (7). Thus, the connections between AMPK activity and metabolic signaling pathways. Alterations in signaling are evident from the changes during HCMV infection have remained unclear. broad changes in cellular phosphorylation that occur during HCMV We confirmed the requirement for AMPK during infection, infection and from the altered activity of multiple kinases. Here we and we show that an AMPK antagonist, compound C, blocks report a comprehensive RNAi screen, which predicts that 106 cellular HCMV-induced changes to glycolysis and inhibits viral gene kinases influence growth of the virus, most of which were not expression. These studies argue that AMPK or a related, com- previously linked to HCMV replication. Multiple elements of the pound C-sensitive kinase is an essential contributor to metabolic AMP-activated protein kinase (AMPK) pathway scored in the screen. changes initiated by HCMV and provide unique insight into As a regulator of carbon and nucleotide metabolism, AMPK is poised potential antiviral strategies. to activate many of the metabolic pathways induced by HCMV infection. An AMPK inhibitor, compound C, blocked a substantial Results portion of HCMV-induced metabolic changes, inhibited the accumu- HumanKinomeScreenIdentifies Putative Effectors of HCMV Replication. lation of all HCMV proteins tested, and markedly reduced the We conducted an siRNA screen of the human kinome to perform an production of infectious progeny. -
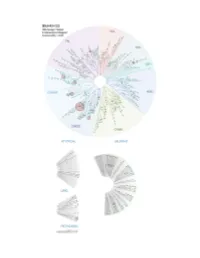
Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) BSJ-03-123 AAK1 AAK1 94 1000 BSJ-03-123 ABL1(E255K)-phosphorylated ABL1 79 1000 BSJ-03-123 ABL1(F317I)-nonphosphorylated ABL1 89 1000 BSJ-03-123 ABL1(F317I)-phosphorylated ABL1 98 1000 BSJ-03-123 ABL1(F317L)-nonphosphorylated ABL1 86 1000 BSJ-03-123 ABL1(F317L)-phosphorylated ABL1 89 1000 BSJ-03-123 ABL1(H396P)-nonphosphorylated ABL1 76 1000 BSJ-03-123 ABL1(H396P)-phosphorylated ABL1 90 1000 BSJ-03-123 ABL1(M351T)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1(Q252H)-nonphosphorylated ABL1 56 1000 BSJ-03-123 ABL1(Q252H)-phosphorylated ABL1 97 1000 BSJ-03-123 ABL1(T315I)-nonphosphorylated ABL1 100 1000 BSJ-03-123 ABL1(T315I)-phosphorylated ABL1 85 1000 BSJ-03-123 ABL1(Y253F)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1-nonphosphorylated ABL1 60 1000 BSJ-03-123 ABL1-phosphorylated ABL1 79 1000 BSJ-03-123 ABL2 ABL2 89 1000 BSJ-03-123 ACVR1 ACVR1 100 1000 BSJ-03-123 ACVR1B ACVR1B 95 1000 BSJ-03-123 ACVR2A ACVR2A 100 1000 BSJ-03-123 ACVR2B ACVR2B 96 1000 BSJ-03-123 ACVRL1 ACVRL1 84 1000 BSJ-03-123 ADCK3 CABC1 90 1000 BSJ-03-123 ADCK4 ADCK4 91 1000 BSJ-03-123 AKT1 AKT1 100 1000 BSJ-03-123 AKT2 AKT2 98 1000 BSJ-03-123 AKT3 AKT3 100 1000 BSJ-03-123 ALK ALK 100 1000 BSJ-03-123 ALK(C1156Y) ALK 78 1000 BSJ-03-123 ALK(L1196M) ALK 100 1000 BSJ-03-123 AMPK-alpha1 PRKAA1 93 1000 BSJ-03-123 AMPK-alpha2 PRKAA2 100 1000 BSJ-03-123 ANKK1 ANKK1 89 1000 BSJ-03-123 ARK5 NUAK1 98 1000 BSJ-03-123 ASK1 MAP3K5 100 1000 BSJ-03-123 ASK2 MAP3K6 92 1000 BSJ-03-123 AURKA -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

De Novo EIF2AK1 and EIF2AK2 Variants Are Associated with Developmental Delay, Leukoencephalopathy, and Neurologic Decompensation
bioRxiv preprint doi: https://doi.org/10.1101/757039; this version posted September 16, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. De novo EIF2AK1 and EIF2AK2 variants are associated with developmental delay, leukoencephalopathy, and neurologic decompensation Dongxue Mao1,2, Chloe M. Reuter3,4, Maura R.Z. Ruzhnikov5,6, Anita E. Beck7, Emily G. Farrow8,9,10, Lisa T. Emrick1,11,12,13, Jill A. Rosenfeld12, Katherine M. Mackenzie5, Laurie Robak2,12,13, Matthew T. Wheeler3,14, Lindsay C. Burrage12,13, Mahim Jain15, Pengfei Liu12, Daniel Calame11,13, Sebastien Küry17,18, Martin Sillesen19, Klaus Schmitz-Abe20, Davide Tonduti21, Luigina Spaccini22, Maria Iascone23, Casie A. Genetti20, Madeline Graf16, Alyssa Tran12, Mercedes Alejandro12, Undiagnosed Diseases Network, Brendan H. Lee12,13, Isabelle Thiffault8,9,24, Pankaj B. Agrawal#,20, Jonathan A. Bernstein#,3,25, Hugo J. Bellen#,2,12,26,27,28, Hsiao- Tuan Chao#,1,2,11,12,13,28,27,29 #Correspondence should be addressed: [email protected] (P.A.), [email protected] (J.A.B.), [email protected] (H.J.B.), [email protected] (H.T.C.) 1Department of Pediatrics, Baylor College of Medicine (BCM), Houston, TX 2Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Houston, TX 3Stanford Center for Undiagnosed Diseases, Stanford University, Stanford, CA 4Stanford Center for Inherited Cardiovascular Disease, Division of Cardiovascular Medicine, -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -
Type of the Paper (Article
Supplementary Materials: Identification of Linkages between EDCs in Personal Care Products and Breast Cancer through Data Integration Combined with Gene Network Analysis Hyeri Jeong 1,2, Jongwoon Kim 1,2,* and Youngjun Kim 1,2 Table S1. Interacting genes and their network types of the 27 common related genes between four selected EDCs and ER positive breast cancer based on GeneMANIA network analysis. Degree Centrality Gene Symbol Official Full Name Interacting Gene Networks * AKT1 2 AR 2, 3, 5, 6 BRCA1 2, 3 CASP8 7 EP300 2, 3 ERBB2 2 13 ESR1 Estrogen receptor 1 HDAC5 2 NCOA1 2, 3 NCOA7 2, 3 PIK3CA 2, 3 SLC10A1 1 SMO 1 TP53 2 AKT1 1 AR 2, 7 BCL6 2, 3 BRCA1 2, 3 CASP8 2 EP300 2, 7 12 TP53 Tumor protein p53 ERBB2 1 ESR1 2 HDAC5 2 MTOR 2 NCOA1 2 SMO 1 AKT1 1, 3 AR 2, 3 BRCA1 2 CYP1A1 1 DUSP10 1, 7 Nuclear receptor EP300 1, 2, 3, 6 12 NCOA1 coactivator 1 ESR1 2, 3 HDAC5 7 KLHL24 1 NCOA7 7 PTCH1 7 TP53 2 AR 2 BRCA1 2 EP300 2 ERBB2 1 ESR1 2 AKT serine/threonine 11 AKT1 MAP2K2 1 kinase 1 MTOR 2, 3, 5, 4 NCOA1 1, 3 PIK3CA 2, 3 SMO 3 TP53 1 Int. J. Environ. Res. Public Health 2017, 14 S2 of S7 Table S1. Cont. Degree Centrality Gene Symbol Official Full Name Interacting Gene Networks * ABCG1 7 APOB 1, 7 EP300 2, 7 GABRR1 7 HDAC5 2, 3 11 BCL6 B-cell CLL/lymphoma 6 KLHL24 1 PIK3CA 1 PTCH1 7 SLC10A1 1 SMO 2 TP53 2, 3 AKT1 2 BRCA1 2, 3 CASP8 1, 3, 4 EP300 2 ESR1 2, 3, 5, 6 10 AR Androgen receptor NCOA1 2, 3 PIK3CA 3 SLC10A1 1 SMO 1 TP53 2, 4 AKT1 2 AR 2 BCL6 2, 7 BRCA1 2, 7 E1A binding protein 9 EP300 ESR1 2, 3 p300 KLHL24 1 NCOA1 1, 2, 3, 6 PIK3CA 1 TP53 2, 7 ABCG1 1 AKT1 2, 3 AR 3 Phosphatidylinositol- BCL6 1 4,5-bisphosphate 9 PIK3CA EP300 1 3-kinase catalytic ERBB2 3 subunit alpha ESR1 2, 3 KLHL24 1 MTOR 3, 6 AR 1 AKT1 3 BCL6 2 Smoothened, frizzled CYP1A1 1 8 SMO class receptor ESR1 1 ERBB2 1 PTCH1 2, 3 TP53 1 AKT1 2 AR 2, 3 EP300 2, 7 BRCA1, DNA 7 BRCA1 ESR1 2, 3 repair associated MTOR 2 NCOA1 2 TP53 2, 3 AR 2, 3, 4 CYP1A1 1 ESR1 7 7 CASP8 Caspase 8 GABRA6 7 KLHL24 7 MTOR 7 TP53 2 Int. -

Silencing of Lncrna H19 Enhances the Sensitivity to X-Rays and Carbon-Ions Through the Mir-130A-3P /WNK3 Signal Axis in Non-Small-Cell Lung Cancer Cells
Silencing of lncRNA H19 Enhances the Sensitivity to X-rays and Carbon-Ions Through the miR-130a-3p /WNK3 Signal Axis in Non-Small-Cell Lung Cancer Cells Xueshan Zhao Lanzhou University First Aliated Hospital https://orcid.org/0000-0002-1194-7617 Xiaodong Jin Institute of Modern Physics Chinese Academy of Sciences Qiuning Zhang Institute of Modern Physics Chinese Academy of Sciences Ruifeng Liu Institute of Modern Physics Chinese Academy of Sciences Hongtao Luo Institute of Modern Physics Chinese Academy of Sciences Zhen Yang Lanzhou University School of Basic Medical Sciences Yichao Geng Lanzhou University First Aliated Hospital Shuangwu Feng Lanzhou University First Aliated Hospital Chengcheng Li Lanzhou University First Aliated Hospital Lina Wang Lanzhou University First Aliated Hospital Xiaohu Wang ( [email protected] ) Lanzhou University First Aliated Hospital Qiang Li Institute of Modern Physics Chinese Academy of Sciences Research Article Keywords: LncRNA H19, MiR-130a-3p, WNK3, Non-small-cell lung cancer, Radiotherapy Posted Date: August 10th, 2021 Page 1/20 DOI: https://doi.org/10.21203/rs.3.rs-768334/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 2/20 Abstract Background: LncRNA H19 was believed to act as an oncogene in various types of tumors and was considered to be a therapeutic target and diagnosis marker. However, the role of lncRNA H19 in regulating the radiosensitivity of non-small cell lung cancer (NSCLC) cells was unknown. However, the effects of lncRNA H19 on radiosensitivity of NSCLC were not clear. Methods: The expression proles of lncRNAs were explored via transcriptome sequencing in NSCLC.