Comparative Transcriptome Profiling of Selected Osmotic Regulatory Proteins in the Gill During Seawater Acclimation of Chum Salm
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

LRRC8A Regulates Hypotonicity-Induced NLRP3 Inflammasome Activation
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.11.146225; this version posted June 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. LRRC8A regulates hypotonicity-induced NLRP3 inflammasome activation Jack P. Green1*,#, Tessa Swanton1*, Lucy V. Morris1, Lina Y. El-Sharkawy2, James Cook1, Shi Yu1, James Beswick2, Antony Adamson3, Neil Humphreys3,4, Richard A. Bryce2, Sally Freeman2, Catherine Lawrence1,5, David Brough1,5,# 1Division of Neuroscience and Experimental Psychology, School of Biological Sciences, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, University of Manchester, AV Hill Building, Oxford Road, Manchester, M13 9PT, UK. 2Division of Pharmacy and Optometry, School of Health Sciences, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, The University of Manchester, Stopford Building, Oxford Road, Manchester, M13 9PT, UK. 3Genome Editing Unit Core Facility, Faculty of Biology, Medicine and Health, University of Manchester, Manchester, UK. 4Present address: EMBL-Rome, Epigenetics and Neurobiology Unit, Adriano Buzzati- Traverso Campus, Via Ramarini 32, 00015 Monterotondo (RM), Italy. 5Lydia Becker Institute of Immunology and Inflammation, University of Manchester, Manchester M13 9PT, UK. * Contributed equally # To whom correspondence should be addressed: David Brough. Tel: +44 (0)161 275 5039; Email: [email protected] Jack Green. Tel +44 (0)161 306 0502; Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.11.146225; this version posted June 12, 2020. -

Trafficking to the Cell Surface of Amino Acid Transporter SLC6A14
cells Article Trafficking to the Cell Surface of Amino Acid Transporter SLC6A14 Upregulated in Cancer Is Controlled by Phosphorylation of SEC24C Protein by AKT Kinase Vasylyna Kovalchuk and Katarzyna A. Nał˛ecz* Nencki Institute of Experimental Biology, Polish Academy of Sciences, 3 Pasteur Street, PL-02-093 Warsaw, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-225892303 Abstract: Cancer cells need a constant supply of nutrients. SLC6A14, an amino acid transporter B0,+ (ATB0,+) that is upregulated in many cancers, transports all but acidic amino acids. In its exit from the endoplasmic reticulum (ER), it is recognized by the SEC24C subunit of coatomer II (COPII) for further vesicular trafficking to the plasma membrane. SEC24C has previously been shown to be phosphorylated by protein kinase B/AKT, which is hyper-activated in cancer; therefore, we analyzed the influence of AKT on SLC6A14 trafficking to the cell surface. Studies on overexpressed and en- dogenous transporters in the breast cancer cell line MCF-7 showed that AKT inhibition with MK-2206 correlated with a transient increase of the transporter in the plasma membrane, not resulting from the inhibition of ER-associated protein degradation. Two-dimensional electrophoresis demonstrated the decreased phosphorylation of SLC6A14 and SEC24C upon AKT inhibition. A proximity ligation assay confirmed this conclusion: AKT inhibition is correlated with decreased SLC6A14 phosphothreonine Citation: Kovalchuk, V.; Nał˛ecz,K.A. Trafficking to the Cell Surface of and SEC24C phosphoserine. Augmented levels of SLC6A14 in plasma membrane led to increased Amino Acid Transporter SLC6A14 leucine transport. These results show that the inactivation of AKT can rescue amino acid delivery Upregulated in Cancer Is Controlled through SLC6A14 trafficking to the cell surface, supporting cancer cell survival. -

GOPC-ROS1 Fusion Due to Microdeletion at 6Q22 Is an Oncogenic Driver in a Subset of Pediatric Gliomas And
J Neuropathol Exp Neurol Vol. 78, No. 12, December 2019, pp. 1089–1099 doi: 10.1093/jnen/nlz093 ORIGINAL ARTICLE GOPC-ROS1 Fusion Due to Microdeletion at 6q22 Is an Oncogenic Driver in a Subset of Pediatric Gliomas and Glioneuronal Tumors Downloaded from https://academic.oup.com/jnen/article/78/12/1089/5593615 by New York University user on 25 August 2020 Timothy E. Richardson, DO, PhD, Karen Tang, MD, Varshini Vasudevaraja, MS, Jonathan Serrano, BS, Christopher M. William, MD, PhD, Kanish Mirchia, MD, Christopher R. Pierson, MD, PhD, Jeffrey R. Leonard, MD, Mohamed S. AbdelBaki, MD, Kathleen M. Schieffer, PhD, Catherine E. Cottrell, PhD, Zulma Tovar-Spinoza, MD, Melanie A. Comito, MD, Daniel R. Boue, MD, PhD, George Jour, MD, and Matija Snuderl, MD features; the third tumor aligned best with glioblastoma and Abstract showed no evidence of neuronal differentiation. Copy number ROS1 is a transmembrane receptor tyrosine kinase proto- profiling revealed chromosome 6q22 microdeletions correspond- oncogene that has been shown to have rearrangements with sev- ingtotheGOPC-ROS1 fusion in all 3 cases and methylation pro- eral genes in glioblastoma and other neoplasms, including intra- filing showed that the tumors did not cluster together as a single chromosomal fusion with GOPC due to microdeletions at 6q22.1. entity or within known methylation classes by t-Distributed Sto- ROS1 fusion events are important findings in these tumors, as chastic Neighbor Embedding. they are potentially targetable alterations with newer tyrosine ki- nase inhibitors; however, whether these tumors represent a dis- Key Words: 6q22, Astrocytoma, Brain tumor, GOPC, Pediatric gli- tinct entity remains unknown. -

Clustering of the Structures of Protein Kinase Activation Loops
bioRxiv preprint doi: https://doi.org/10.1101/395723; this version posted August 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Clustering of the structures of protein kinase activation loops: A new nomenclature for active and inactive kinase structures Vivek Modi Roland L. Dunbrack, Jr.* Institute for Cancer Research Fox Chase Cancer Center Philadelphia PA 19111 *[email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/395723; this version posted August 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract TarGeting protein kinases is an important strateGy for intervention in cancer. Inhibitors are directed at the conserved active conformation or a variety of inactive conformations. While attempts have been made to classify these conformations, a structurally rigorous cataloGue of states has not been achieved. The kinase activation loop is crucial for catalysis and beGins with the conserved DFGmotif (Asp-Phe-Gly). This motif is observed in two major classes of conformations, DFGin - an ensemble of active and inactive conformations where the Phe residue is in contact with the C-helix of the N-terminal lobe, and DFGout - an inactive form where Phe occupies the ATP site exposinG the C-helix pocket. -

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Label-Free Proteomic Methodology for the Analysis of Human Kidney Stone Matrix Composition Frank A
Witzmann et al. Proteome Science (2016) 14:4 DOI 10.1186/s12953-016-0093-x METHODOLOGY Open Access Label-free proteomic methodology for the analysis of human kidney stone matrix composition Frank A. Witzmann1*, Andrew P. Evan2, Fredric L. Coe3, Elaine M. Worcester3, James E. Lingeman4 and James C. Williams Jr2 Abstract Background: Kidney stone matrix protein composition is an important yet poorly understood aspect of nephrolithiasis. We hypothesized that this proteome is considerably more complex than previous reports have indicated and that comprehensive proteomic profiling of the kidney stone matrix may demonstrate relevant constitutive differences between stones. We have analyzed the matrices of two unique human calcium oxalate stones (CaOx-Ia and CaOx-Id) using a simple but effective chaotropic reducing solution for extraction/solubilization combined with label-free quantitative mass spectrometry to generate a comprehensive profile of their proteomes, including physicochemical and bioinformatic analysis.` Results: We identified and quantified 1,059 unique protein database entries in the two human kidney stone samples, revealing a more complex proteome than previously reported. Protein composition reflects a common range of proteins related to immune response, inflammation, injury, and tissue repair, along with a more diverse set of proteins unique to each stone. Conclusion: The use of a simple chaotropic reducing solution and moderate sonication for extraction and solubilization of kidney stone powders combined with label-free quantitative mass spectrometry has yielded the most comprehensive list to date of the proteins that constitute the human kidney stone proteome. Keywords: Calcium oxalate, Kidney stone, Label-free quantitative liquid chromatography–tandem mass spectrometry, Matrix protein, Nephrolithiasis, Proteomics Background deposition is the primary event, at least in the formation The organic matrix within urinary stones has long been of CaOx stones over plaque. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The LRRC8A:C Heteromeric Channel Is a Cgamp Transporter and the Dominant
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.13.948273; this version posted February 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The LRRC8A:C Heteromeric Channel Is a cGAMP Transporter and the Dominant cGAMP Importer in Human Vasculature Cells Lauren J. Lahey1,2, Xianlan Wen3, Rachel E. Mardjuki2,4, Volker Böhnert2,5, Christopher Ritchie2,5, Jacqueline A. Carozza2,4, Gaelen T. Hess2,6, Merritt Maduke3, Michael C. Bassik2,6, and Lingyin Li2,5,7,* 1Biophysics Program, 2Stanford ChEM-H, 3Department of Molecular and Cellular Physiology, 4Department of Chemistry, 5Department of Biochemistry, 6Department of Genetics, Stanford University School of Medicine, Stanford, CA 94305 USA. 7Lead Contact *Correspondence: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.02.13.948273; this version posted February 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. SUMMARY Extracellular 2'3'-cyclic-GMP-AMP (cGAMP) is an immunotransmitter secreted by cancer cells and taken up by host cells to activate the anti-cancer STING pathway. No cGAMP exporter has been identified, and SLC19A1, a recently identified cGAMP importer, does not account for the import actiVity in most cell types. Here, we identify the LRRC8A:C heteromeric channel, a volume-regulated anion channel (VRAC), as a cGAMP transporter. This channel mediates cGAMP import or export depending on the cGAMP chemical gradient. cGAMP influences anion influx through VRAC, indicating it is likely a direct substrate of the channel. -

Russell Doolittle Biochemistry Laboratory Records
http://oac.cdlib.org/findaid/ark:/13030/tf0v19n7xm No online items Russell Doolittle Biochemistry Laboratory Records Special Collections & Archives, UC San Diego Special Collections & Archives, UC San Diego Copyright 2005 9500 Gilman Drive La Jolla 92093-0175 [email protected] URL: http://libraries.ucsd.edu/collections/sca/index.html Russell Doolittle Biochemistry MSS 0077 1 Laboratory Records Descriptive Summary Languages: English Contributing Institution: Special Collections & Archives, UC San Diego 9500 Gilman Drive La Jolla 92093-0175 Title: Russell Doolittle Biochemistry Laboratory Records Identifier/Call Number: MSS 0077 Physical Description: 87.4 Linear feet (83 records cartons, 8 archives boxes, 16 oversize folders and 1 art bin item) Date (inclusive): 1971 - 1998 Abstract: The records (1971-1998) of Dr. Russell F. Doolittle's biochemistry laboratory at the University of California, San Diego include notebooks related to the first determination of the complete sequence of amino acids in the human fibrinogen molecule, paper files for the amino acid sequences contained in the protein sequence data bank called NEWAT, as well as other research, correspondence and Protein Society files. Preferred Citation Russell Doolittle Biochemistry Laboratory Records, MSS 0077. Special Collections & Archives, UC San Diego. Administrative History Dr. Russell F. Doolittle, professor of chemistry at the University of California, San Diego, headed a campus science laboratory that conducts research in the evolutionary and structural aspects of proteins. In 1979, Doolittle's laboratory successfully analyzed the structure of the amino acid sequence for the human fibrinogen molecule. During that study, ten to twenty postdoctoral, graduate and undergraduate researchers worked to pull apart and analyze the amino acid sequences in the alpha, beta and gamma chains of fibrinogen. -
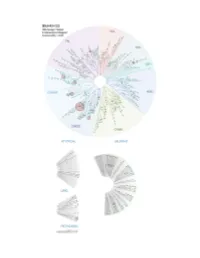
Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) BSJ-03-123 AAK1 AAK1 94 1000 BSJ-03-123 ABL1(E255K)-phosphorylated ABL1 79 1000 BSJ-03-123 ABL1(F317I)-nonphosphorylated ABL1 89 1000 BSJ-03-123 ABL1(F317I)-phosphorylated ABL1 98 1000 BSJ-03-123 ABL1(F317L)-nonphosphorylated ABL1 86 1000 BSJ-03-123 ABL1(F317L)-phosphorylated ABL1 89 1000 BSJ-03-123 ABL1(H396P)-nonphosphorylated ABL1 76 1000 BSJ-03-123 ABL1(H396P)-phosphorylated ABL1 90 1000 BSJ-03-123 ABL1(M351T)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1(Q252H)-nonphosphorylated ABL1 56 1000 BSJ-03-123 ABL1(Q252H)-phosphorylated ABL1 97 1000 BSJ-03-123 ABL1(T315I)-nonphosphorylated ABL1 100 1000 BSJ-03-123 ABL1(T315I)-phosphorylated ABL1 85 1000 BSJ-03-123 ABL1(Y253F)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1-nonphosphorylated ABL1 60 1000 BSJ-03-123 ABL1-phosphorylated ABL1 79 1000 BSJ-03-123 ABL2 ABL2 89 1000 BSJ-03-123 ACVR1 ACVR1 100 1000 BSJ-03-123 ACVR1B ACVR1B 95 1000 BSJ-03-123 ACVR2A ACVR2A 100 1000 BSJ-03-123 ACVR2B ACVR2B 96 1000 BSJ-03-123 ACVRL1 ACVRL1 84 1000 BSJ-03-123 ADCK3 CABC1 90 1000 BSJ-03-123 ADCK4 ADCK4 91 1000 BSJ-03-123 AKT1 AKT1 100 1000 BSJ-03-123 AKT2 AKT2 98 1000 BSJ-03-123 AKT3 AKT3 100 1000 BSJ-03-123 ALK ALK 100 1000 BSJ-03-123 ALK(C1156Y) ALK 78 1000 BSJ-03-123 ALK(L1196M) ALK 100 1000 BSJ-03-123 AMPK-alpha1 PRKAA1 93 1000 BSJ-03-123 AMPK-alpha2 PRKAA2 100 1000 BSJ-03-123 ANKK1 ANKK1 89 1000 BSJ-03-123 ARK5 NUAK1 98 1000 BSJ-03-123 ASK1 MAP3K5 100 1000 BSJ-03-123 ASK2 MAP3K6 92 1000 BSJ-03-123 AURKA -

Selective Transport of Neurotransmitters and Modulators by Distinct Volume-Regulated LRRC8 Anion Channels Darius Lutter1,2,3, Florian Ullrich1,2, Jennifer C
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 1122-1133 doi:10.1242/jcs.196253 RESEARCH ARTICLE Selective transport of neurotransmitters and modulators by distinct volume-regulated LRRC8 anion channels Darius Lutter1,2,3, Florian Ullrich1,2, Jennifer C. Lueck1,2,3, Stefan Kempa2 and Thomas J. Jentsch1,2,4,* ABSTRACT volume regulation (Jentsch, 2016; Nilius et al., 1997; Pedersen In response to swelling, mammalian cells release chloride and et al., 2016). This plasma membrane channel, which appears to be organic osmolytes through volume-regulated anion channels expressed in all cells, is normally closed but can be activated by cell swelling or other cellular signaling cascades. In most cells, VRAC (VRACs). VRACs are heteromers of LRRC8A and other LRRC8 − isoforms (LRRC8B to LRRC8E), which are co-expressed in HEK293 opening leads to an efflux of Cl , which, together with an efflux of + + and most other cells. The spectrum of VRAC substrates and its K through independent K channels, lowers the intracellular dependence on particular LRRC8 isoforms remains largely unknown. osmolarity and, by secondarily driving water efflux, results in We show that, besides the osmolytes taurine and myo-inositol, regulatory volume decrease. Cell swelling also stimulates an efflux ‘ ’ myo LRRC8 channels transport the neurotransmitters glutamate, of organic osmolytes such as -inositol, taurine and glutamate. aspartate and γ-aminobutyric acid (GABA) and the co-activator It was controversial whether this release occurs through VRAC or D-serine. HEK293 cells engineered to express defined subsets of through a distinct channel named VSOAC (for volume-sensitive LRRC8 isoforms were used to elucidate the subunit-dependence organic osmolyte/anion channel) (Jackson and Strange, 1993; of transport. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia.