Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Sulfite Dehydrogenases in Organotrophic Bacteria : Enzymes
Sulfite dehydrogenases in organotrophic bacteria: enzymes, genes and regulation. Dissertation zur Erlangung des akademischen Grades des Doktors der Naturwissenschaften (Dr. rer. nat.) an der Universität Konstanz Fachbereich Biologie vorgelegt von Sabine Lehmann Tag der mündlichen Prüfung: 10. April 2013 1. Referent: Prof. Dr. Bernhard Schink 2. Referent: Prof. Dr. Andrew W. B. Johnston So eine Arbeit wird eigentlich nie fertig, man muss sie für fertig erklären, wenn man nach Zeit und Umständen das möglichste getan hat. (Johann Wolfgang von Goethe, Italienische Reise, 1787) DANKSAGUNG An dieser Stelle möchte ich mich herzlich bei folgenden Personen bedanken: . Prof. Dr. Alasdair M. Cook (Universität Konstanz, Deutschland), der mir dieses Thema und seine Laboratorien zur Verfügung stellte, . Prof. Dr. Bernhard Schink (Universität Konstanz, Deutschland), für seine spontane und engagierte Übernahme der Betreuung, . Prof. Dr. Andrew W. B. Johnston (University of East Anglia, UK), für seine herzliche und bereitwillige Aufnahme in seiner Arbeitsgruppe, seiner engagierten Unter- stützung, sowie für die Übernahme des Koreferates, . Prof. Dr. Frithjof C. Küpper (University of Aberdeen, UK), für seine große Hilfsbereitschaft bei der vorliegenden Arbeit und geplanter Manuskripte, als auch für die mentale Unterstützung während der letzten Jahre! Desweiteren möchte ich herzlichst Dr. David Schleheck für die Übernahme des Koreferates der mündlichen Prüfung sowie Prof. Dr. Alexander Bürkle, für die Übernahme des Prüfungsvorsitzes sowie für seine vielen hilfreichen Ratschläge danken! Ein herzliches Dankeschön geht an alle beteiligten Arbeitsgruppen der Universität Konstanz, der UEA und des SAMS, ganz besonders möchte ich dabei folgenden Personen danken: . Dr. David Schleheck und Karin Denger, für die kritische Durchsicht dieser Arbeit, der durch und durch sehr engagierten Hilfsbereitschaft bei Problemen, den zahlreichen wissenschaftlichen Diskussionen und für die aufbauenden Worte, . -

Advances in Chitin/Chitosan Characterization and Applications
Advances in Chitin/Chitosan Characterization and Applications Edited by Marguerite Rinaudo and Francisco M. Goycoolea Printed Edition of the Special Issue Published in Polymers www.mdpi.com/journal/polymers Advances in Chitin/Chitosan Characterization and Applications Advances in Chitin/Chitosan Characterization and Applications Special Issue Editors Marguerite Rinaudo Francisco M. Goycoolea MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade Special Issue Editors Marguerite Rinaudo Francisco M. Goycoolea University of Grenoble Alpes University of Leeds France UK Editorial Office MDPI St. Alban-Anlage 66 4052 Basel, Switzerland This is a reprint of articles from the Special Issue published online in the open access journal Polymers (ISSN 2073-4360) from 2017 to 2018 (available at: https://www.mdpi.com/journal/polymers/ special issues/chitin chitosan) For citation purposes, cite each article independently as indicated on the article page online and as indicated below: LastName, A.A.; LastName, B.B.; LastName, C.C. Article Title. Journal Name Year, Article Number, Page Range. ISBN 978-3-03897-802-2 (Pbk) ISBN 978-3-03897-803-9 (PDF) c 2019 by the authors. Articles in this book are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book as a whole is distributed by MDPI under the terms and conditions of the Creative Commons license CC BY-NC-ND. Contents About the Special Issue Editors ..................................... ix Preface to ”Advances in Chitin/Chitosan Characterization and Applications” ......... -

(12) Patent Application Publication (10) Pub. No.: US 2014/0155567 A1 Burk Et Al
US 2014O155567A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2014/0155567 A1 Burk et al. (43) Pub. Date: Jun. 5, 2014 (54) MICROORGANISMS AND METHODS FOR (60) Provisional application No. 61/331,812, filed on May THE BIOSYNTHESIS OF BUTADENE 5, 2010. (71) Applicant: Genomatica, Inc., San Diego, CA (US) Publication Classification (72) Inventors: Mark J. Burk, San Diego, CA (US); (51) Int. Cl. Anthony P. Burgard, Bellefonte, PA CI2P 5/02 (2006.01) (US); Jun Sun, San Diego, CA (US); CSF 36/06 (2006.01) Robin E. Osterhout, San Diego, CA CD7C II/6 (2006.01) (US); Priti Pharkya, San Diego, CA (52) U.S. Cl. (US) CPC ................. CI2P5/026 (2013.01); C07C II/I6 (2013.01); C08F 136/06 (2013.01) (73) Assignee: Genomatica, Inc., San Diego, CA (US) USPC ... 526/335; 435/252.3:435/167; 435/254.2: (21) Appl. No.: 14/059,131 435/254.11: 435/252.33: 435/254.21:585/16 (22) Filed: Oct. 21, 2013 (57) ABSTRACT O O The invention provides non-naturally occurring microbial Related U.S. Application Data organisms having a butadiene pathway. The invention addi (63) Continuation of application No. 13/101,046, filed on tionally provides methods of using Such organisms to produce May 4, 2011, now Pat. No. 8,580,543. butadiene. Patent Application Publication Jun. 5, 2014 Sheet 1 of 4 US 2014/O155567 A1 ?ueudos!SMS |?un61– Patent Application Publication Jun. 5, 2014 Sheet 2 of 4 US 2014/O155567 A1 VOJ OO O Z?un61– Patent Application Publication US 2014/O155567 A1 {}}} Hººso Patent Application Publication Jun. -

1471-2164-15-164.Pdf
Meinhardt et al. BMC Genomics 2014, 15:164 http://www.biomedcentral.com/1471-2164/15/164 RESEARCH ARTICLE Open Access Genome and secretome analysis of the hemibiotrophic fungal pathogen, Moniliophthora roreri, which causes frosty pod rot disease of cacao: mechanisms of the biotrophic and necrotrophic phases Lyndel W Meinhardt1*, Gustavo Gilson Lacerda Costa2, Daniela PT Thomazella3, Paulo José PL Teixeira3, Marcelo Falsarella Carazzolle2, Stephan C Schuster4, John E Carlson5, Mark J Guiltinan6, Piotr Mieczkowski7, Andrew Farmer8, Thiruvarangan Ramaraj8, Jayne Crozier9, Robert E Davis10, Jonathan Shao10, Rachel L Melnick1, Gonçalo AG Pereira3 and Bryan A Bailey1 Abstract Background: The basidiomycete Moniliophthora roreri is the causal agent of Frosty pod rot (FPR) disease of cacao (Theobroma cacao), the source of chocolate, and FPR is one of the most destructive diseases of this important perennial crop in the Americas. This hemibiotroph infects only cacao pods and has an extended biotrophic phase lasting up to sixty days, culminating in plant necrosis and sporulation of the fungus without the formation of a basidiocarp. Results: We sequenced and assembled 52.3 Mb into 3,298 contigs that represent the M. roreri genome. Of the 17,920 predicted open reading frames (OFRs), 13,760 were validated by RNA-Seq. Using read count data from RNA sequencing of cacao pods at 30 and 60 days post infection, differential gene expression was estimated for the biotrophic and necrotrophic phases of this plant-pathogen interaction. The sequencing data were used to develop a genome based secretome for the infected pods. Of the 1,535 genes encoding putative secreted proteins, 1,355 were expressed in the biotrophic and necrotrophic phases. -

Pyruvate-Phosphate Dikinase of Oxymonads and Parabasalia and the Evolution of Pyrophosphate-Dependent Glycolysis in Anaerobic Eukaryotes† Claudio H
EUKARYOTIC CELL, Jan. 2006, p. 148–154 Vol. 5, No. 1 1535-9778/06/$08.00ϩ0 doi:10.1128/EC.5.1.148–154.2006 Copyright © 2006, American Society for Microbiology. All Rights Reserved. Pyruvate-Phosphate Dikinase of Oxymonads and Parabasalia and the Evolution of Pyrophosphate-Dependent Glycolysis in Anaerobic Eukaryotes† Claudio H. Slamovits and Patrick J. Keeling* Canadian Institute for Advanced Research, Botany Department, University of British Columbia, 3529-6270 University Boulevard, Vancouver, British Columbia V6T 1Z4, Canada Received 29 September 2005/Accepted 8 November 2005 In pyrophosphate-dependent glycolysis, the ATP/ADP-dependent enzymes phosphofructokinase (PFK) and pyruvate kinase are replaced by the pyrophosphate-dependent PFK and pyruvate phosphate dikinase (PPDK), respectively. This variant of glycolysis is widespread among bacteria, but it also occurs in a few parasitic anaerobic eukaryotes such as Giardia and Entamoeba spp. We sequenced two genes for PPDK from the amitochondriate oxymonad Streblomastix strix and found evidence for PPDK in Trichomonas vaginalis and other parabasalia, where this enzyme was thought to be absent. The Streblomastix and Giardia genes may be related to one another, but those of Entamoeba and perhaps Trichomonas are distinct and more closely related to bacterial homologues. These findings suggest that pyrophosphate-dependent glycolysis is more widespread in eukaryotes than previously thought, enzymes from the pathway coexists with ATP-dependent more often than previously thought and may be spread by lateral transfer of genes for pyrophosphate-dependent enzymes from bacteria. Adaptation to anaerobic metabolism is a complex process (PPDK), respectively (for a comparison of these reactions, see involving changes to many proteins and pathways of critical reference 21). -

Characterization of a Eukaryotic Type Serine/Threonine Protein Kinase And
Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates Linda Nova´ kova´ 1, Lenka Saskova´ 1, Petra Pallova´ 1, Jirˇı´ Janecˇek1, Jana Novotna´ 1, Alesˇ Ulrych1, Jose Echenique2, Marie-Claude Trombe3 and Pavel Branny1 1 Cell and Molecular Microbiology Division, Institute of Microbiology, Czech Academy of Sciences, Prague, Czech Republic 2 Departamento de Bioquı´mica Clı´nica, Facultad de Ciencias Quı´micas, Universidad Nacional de Co´ rdoba, Medina Allende esq Haya de la Torre, Ciudad Universitaria, Co´ rdoba, Argentina 3 Centre Hospitalo-Universitaire de Rangueil, Universite´ Paul Sabatier, Toulouse, France Keywords Searching the genome sequence of Streptococcus pneumoniae revealed the phosphoglucosamine mutase; presence of a single Ser ⁄ Thr protein kinase gene stkP linked to protein phosphoproteome; protein phosphatase; phosphatase phpP. Biochemical studies performed with recombinant StkP serine ⁄ threonine protein kinase; suggest that this protein is a functional eukaryotic-type Ser ⁄ Thr protein Streptococcus pneumoniae kinase. In vitro kinase assays and Western blots of S. pneumoniae subcellu- Correspondence lar fractions revealed that StkP is a membrane protein. PhpP is a soluble P. Branny, Cell and Molecular Microbiology protein with manganese-dependent phosphatase activity in vitro against a Division, Institute of Microbiology, Czech synthetic substrate RRA(pT)VA. Mutations in the invariant aspartate resi- Academy of Sciences, Vı´denˇ ska´ 1083, dues implicated in the metal binding completely abolished PhpP activity. 142 20 Prague 4, Czech Republic Autophosphorylated form of StkP was shown to be a substrate for PhpP. Fax: +420 2 41722257 These results suggest that StkP and PhpP could operate as a functional Tel: +420 2 41062658 E-mail: [email protected] pair in vivo. -

Changes in the Sclerotinia Sclerotiorum Transcriptome During Infection of Brassica Napus
Seifbarghi et al. BMC Genomics (2017) 18:266 DOI 10.1186/s12864-017-3642-5 RESEARCHARTICLE Open Access Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus Shirin Seifbarghi1,2, M. Hossein Borhan1, Yangdou Wei2, Cathy Coutu1, Stephen J. Robinson1 and Dwayne D. Hegedus1,3* Abstract Background: Sclerotinia sclerotiorum causes stem rot in Brassica napus, which leads to lodging and severe yield losses. Although recent studies have explored significant progress in the characterization of individual S. sclerotiorum pathogenicity factors, a gap exists in profiling gene expression throughout the course of S. sclerotiorum infection on a host plant. In this study, RNA-Seq analysis was performed with focus on the events occurring through the early (1 h) to the middle (48 h) stages of infection. Results: Transcript analysis revealed the temporal pattern and amplitude of the deployment of genes associated with aspects of pathogenicity or virulence during the course of S. sclerotiorum infection on Brassica napus. These genes were categorized into eight functional groups: hydrolytic enzymes, secondary metabolites, detoxification, signaling, development, secreted effectors, oxalic acid and reactive oxygen species production. The induction patterns of nearly all of these genes agreed with their predicted functions. Principal component analysis delineated gene expression patterns that signified transitions between pathogenic phases, namely host penetration, ramification and necrotic stages, and provided evidence for the occurrence of a brief biotrophic phase soon after host penetration. Conclusions: The current observations support the notion that S. sclerotiorum deploys an array of factors and complex strategies to facilitate host colonization and mitigate host defenses. This investigation provides a broad overview of the sequential expression of virulence/pathogenicity-associated genes during infection of B. -

Table 6. Putative Genes Involved in the Utilization of Carbohydrates in G
Table 6. Putative genes involved in the utilization of carbohydrates in G. thermodenitrificans NG80-2 genome Carbohydrates* Enzymes Gene ID Glycerol Glycerol Kinase GT1216 Glycerol-3-phosphate dehydrogenase, aerobic GT2089 NAD(P)H-dependent glycerol-3-phosphate dehydrogenase GT2153 Enolase GT3003 2,3-bisphosphoglycerate-independentphosphoglycerate mutase GT3004 Triosephosphate isomerase GT3005 3-phosphoglycerate kinase GT3006 Glyceraldehyde-3-phosphate dehydrogenase GT3007 Phosphoglycerate mutase GT1326 Pyruvate kinase GT2663 L-Arabinose L-arabinose isomerase GT1795 L-ribulokinase GT1796 L-ribulose 5-phosphate 4-epimerase GT1797 D-Ribose Ribokinase GT3174 Transketolase GT1187 Ribose 5-phosphate epimerase GT3316 D-Xylose Xylose kinase GT1756 Xylose isomerase GT1757 D-Galactose Galactokinase GT2086 Galactose-1-phosphate uridyltransferase GT2084 UDP-glucose 4-epimerase GT2085 Carbohydrates* Enzymes Gene ID D-Fructose 1-phosphofructokinase GT1727 Fructose-1,6-bisphosphate aldolase GT1805 Fructose-1,6-bisphosphate aldolase type II GT3331 Triosephosphate isomerase GT3005 D-Mannose Mannnose-6 phospate isomelase GT3398 6-phospho-1-fructokinase GT2664 D-Mannitol Mannitol-1-phosphate dehydrogenase GT1844 N-Acetylglucosamine N-acetylglucosamine-6-phosphate deacetylase GT2205 N-acetylglucosamine-6-phosphate isomerase GT2204 D-Maltose Alpha-1,4-glucosidase GT0528, GT1643 Sucrose Sucrose phosphorylase GT3215 D-Trehalose Alpha-glucosidase GT1643 Glucose kinase GT2381 Inositol Myo-inositol catabolism protein iolC;5-dehydro-2- GT1807 deoxygluconokinase -

Dissimilation of Cysteate Via 3-Sulfolactate Sulfo-Lyase and a Sulfate Exporter in Paracoccus Pantotrophus NKNCYSA
Microbiology (2005), 151, 737–747 DOI 10.1099/mic.0.27548-0 Dissimilation of cysteate via 3-sulfolactate sulfo-lyase and a sulfate exporter in Paracoccus pantotrophus NKNCYSA Ulrike Rein,1 Ronnie Gueta,1 Karin Denger,1 Ju¨rgen Ruff,1 Klaus Hollemeyer2 and Alasdair M. Cook1 Correspondence 1Department of Biology, The University, D-78457 Konstanz, Germany Alasdair Cook 2Institute of Biochemical Engineering, Saarland University, Box 50 11 50, D-66041 [email protected] Saarbru¨cken, Germany Paracoccus pantotrophus NKNCYSA utilizes (R)-cysteate (2-amino-3-sulfopropionate) as a sole source of carbon and energy for growth, with either nitrate or molecular oxygen as terminal electron acceptor, and the specific utilization rate of cysteate is about 2 mkat (kg protein)”1. The initial degradative reaction is catalysed by an (R)-cysteate : 2-oxoglutarate aminotransferase, which yields 3-sulfopyruvate. The latter was reduced to 3-sulfolactate by an NAD-linked sulfolactate dehydrogenase [3?3 mkat (kg protein)”1]. The inducible desulfonation reaction was not detected initially in cell extracts. However, a strongly induced protein with subunits of 8 kDa (a) and 42 kDa (b) was found and purified. The corresponding genes had similarities to those encoding altronate dehydratases, which often require iron for activity. The purified enzyme could then be shown to convert 3-sulfolactate to sulfite and pyruvate and it was termed sulfolactate sulfo-lyase (Suy). A high level of sulfite dehydrogenase was also induced during growth with cysteate, and the organism excreted sulfate. A putative regulator, OrfR, was encoded upstream of suyAB on the reverse strand. Downstream of suyAB was suyZ, which was cotranscribed with suyB. -

Evolving a New Efficient Mode of Fructose Utilization For
fbioe-09-669093 May 22, 2021 Time: 22:55 # 1 ORIGINAL RESEARCH published: 28 May 2021 doi: 10.3389/fbioe.2021.669093 Evolving a New Efficient Mode of Fructose Utilization for Improved Bioproduction in Corynebacterium glutamicum Irene Krahn1, Daniel Bonder2, Lucía Torregrosa-Barragán2, Dominik Stoppel1, 1 3 1 3,4 Edited by: Jens P. Krause , Natalie Rosenfeldt , Tobias M. Meiswinkel , Gerd M. Seibold , Pablo Ivan Nikel, Volker F. Wendisch1 and Steffen N. Lindner1,2* Novo Nordisk Foundation Center 1 2 for Biosustainability (DTU Biosustain), Chair of Genetics of Prokaryotes, Faculty of Biology and CeBiTec, Bielefeld University, Bielefeld, Germany, Systems 3 Denmark and Synthetic Metabolism, Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm, Germany, Institute of Biochemistry, University of Cologne, Cologne, Germany, 4 Department of Biotechnology and Biomedicine, Technical Reviewed by: University of Denmark, Lyngby, Denmark Stephan Noack, Julich-Forschungszentrum, Helmholtz-Verband Deutscher Fructose utilization in Corynebacterium glutamicum starts with its uptake and Forschungszentren (HZ), Germany concomitant phosphorylation via the phosphotransferase system (PTS) to yield Fabien Létisse, UMR 5504 Laboratoire d’Ingénierie intracellular fructose 1-phosphate, which enters glycolysis upon ATP-dependent des Systèmes Biologiques et des phosphorylation to fructose 1,6-bisphosphate by 1-phosphofructokinase. This is known Procédés (LISBP), France to result in a significantly reduced oxidative pentose phosphate pathway (oxPPP) flux *Correspondence: ∼ ∼ Steffen N. Lindner on fructose ( 10%) compared to glucose ( 60%). Consequently, the biosynthesis of [email protected] NADPH demanding products, e.g., L-lysine, by C. glutamicum is largely decreased when fructose is the only carbon source. Previous works reported that fructose Specialty section: This article was submitted to is partially utilized via the glucose-specific PTS presumably generating fructose 6- Synthetic Biology, phosphate. -
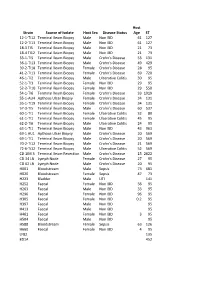
Strain Source of Isolate Host Sex Disease Status Host Age ST
Host Strain Source of Isolate Host Sex Disease Status Age ST 12-1-TI12 Terminal Ileum Biopsy Male Non IBD 61 127 12-2-TI13 Terminal Ileum Biopsy Male Non IBD 61 127 18-3 TI5 Terminal Ileum Biopsy Male Non IBD 21 73 18-4 TI12 Terminal Ileum Biopsy Male Non IBD 21 73 33-1-TI5 Terminal Ileum Biopsy Male Crohn's Disease 53 131 36-1-TI13 Terminal Ileum Biopsy Male Crohn's Disease 49 429 39-2-TI18 Terminal Ileum Biopsy Female Crohn's Disease 28 95 41-2-TI13 Terminal Ileum Biopsy Female Crohn's Disease 69 720 46-1-TI2 Terminal Ileum Biopsy Male Ulcerative Colitis 30 95 52-1-TI3 Terminal Ileum Biopsy Female Non IBD 29 95 52-2-TI10 Terminal Ileum Biopsy Female Non IBD 29 550 54-1-TI6 Terminal Ileum Biopsy Female Crohn's Disease 33 1919 55-1-AU4 Apthous Ulcer Biopsy Female Crohn's Disease 34 131 55-1-TI19 Terminal Ileum Biopsy Female Crohn's Disease 34 131 57-3-TI5 Terminal Ileum Biopsy Male Crohn's Disease 60 537 60-1-TI1 Terminal Ileum Biopsy Female Ulcerative Colitis 32 80 61-1-TI1 Terminal Ileum Biopsy Female Ulcerative Colitis 45 95 62-2-TI6 Terminal Ileum Biopsy Male Ulcerative Colitis 24 95 63-1-TI1 Terminal Ileum Biopsy Male Non IBD 43 963 69-1 AU1 Apthous Ulcer Biopsy Male Crohn's Disease 20 569 69-1-TI1 Terminal Ileum Biopsy Male Crohn's Disease 20 569 70-2-TI12 Terminal Ileum Biopsy Male Crohn's Disease 21 569 72-6-Ti12 Terminal Ileum Biopsy Male Ulcerative Colitis 52 569 CD 1IM 3 Terminal Ileum Resection Male Crohn's Disease 15 2622 CD 34 LN Lymph Node Female Crohn's Disease 27 95 CD 62 LN Lymph Node Male Crohn's Disease 20 95 H001