Within-Host Diversity of Multidrug-Resistant Escherichia Coli in the Gut and Bladder Sofiya G. Shevchenko a Dissertation Submitt
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sulfite Dehydrogenases in Organotrophic Bacteria : Enzymes
Sulfite dehydrogenases in organotrophic bacteria: enzymes, genes and regulation. Dissertation zur Erlangung des akademischen Grades des Doktors der Naturwissenschaften (Dr. rer. nat.) an der Universität Konstanz Fachbereich Biologie vorgelegt von Sabine Lehmann Tag der mündlichen Prüfung: 10. April 2013 1. Referent: Prof. Dr. Bernhard Schink 2. Referent: Prof. Dr. Andrew W. B. Johnston So eine Arbeit wird eigentlich nie fertig, man muss sie für fertig erklären, wenn man nach Zeit und Umständen das möglichste getan hat. (Johann Wolfgang von Goethe, Italienische Reise, 1787) DANKSAGUNG An dieser Stelle möchte ich mich herzlich bei folgenden Personen bedanken: . Prof. Dr. Alasdair M. Cook (Universität Konstanz, Deutschland), der mir dieses Thema und seine Laboratorien zur Verfügung stellte, . Prof. Dr. Bernhard Schink (Universität Konstanz, Deutschland), für seine spontane und engagierte Übernahme der Betreuung, . Prof. Dr. Andrew W. B. Johnston (University of East Anglia, UK), für seine herzliche und bereitwillige Aufnahme in seiner Arbeitsgruppe, seiner engagierten Unter- stützung, sowie für die Übernahme des Koreferates, . Prof. Dr. Frithjof C. Küpper (University of Aberdeen, UK), für seine große Hilfsbereitschaft bei der vorliegenden Arbeit und geplanter Manuskripte, als auch für die mentale Unterstützung während der letzten Jahre! Desweiteren möchte ich herzlichst Dr. David Schleheck für die Übernahme des Koreferates der mündlichen Prüfung sowie Prof. Dr. Alexander Bürkle, für die Übernahme des Prüfungsvorsitzes sowie für seine vielen hilfreichen Ratschläge danken! Ein herzliches Dankeschön geht an alle beteiligten Arbeitsgruppen der Universität Konstanz, der UEA und des SAMS, ganz besonders möchte ich dabei folgenden Personen danken: . Dr. David Schleheck und Karin Denger, für die kritische Durchsicht dieser Arbeit, der durch und durch sehr engagierten Hilfsbereitschaft bei Problemen, den zahlreichen wissenschaftlichen Diskussionen und für die aufbauenden Worte, . -

Dissimilation of Cysteate Via 3-Sulfolactate Sulfo-Lyase and a Sulfate Exporter in Paracoccus Pantotrophus NKNCYSA
Microbiology (2005), 151, 737–747 DOI 10.1099/mic.0.27548-0 Dissimilation of cysteate via 3-sulfolactate sulfo-lyase and a sulfate exporter in Paracoccus pantotrophus NKNCYSA Ulrike Rein,1 Ronnie Gueta,1 Karin Denger,1 Ju¨rgen Ruff,1 Klaus Hollemeyer2 and Alasdair M. Cook1 Correspondence 1Department of Biology, The University, D-78457 Konstanz, Germany Alasdair Cook 2Institute of Biochemical Engineering, Saarland University, Box 50 11 50, D-66041 [email protected] Saarbru¨cken, Germany Paracoccus pantotrophus NKNCYSA utilizes (R)-cysteate (2-amino-3-sulfopropionate) as a sole source of carbon and energy for growth, with either nitrate or molecular oxygen as terminal electron acceptor, and the specific utilization rate of cysteate is about 2 mkat (kg protein)”1. The initial degradative reaction is catalysed by an (R)-cysteate : 2-oxoglutarate aminotransferase, which yields 3-sulfopyruvate. The latter was reduced to 3-sulfolactate by an NAD-linked sulfolactate dehydrogenase [3?3 mkat (kg protein)”1]. The inducible desulfonation reaction was not detected initially in cell extracts. However, a strongly induced protein with subunits of 8 kDa (a) and 42 kDa (b) was found and purified. The corresponding genes had similarities to those encoding altronate dehydratases, which often require iron for activity. The purified enzyme could then be shown to convert 3-sulfolactate to sulfite and pyruvate and it was termed sulfolactate sulfo-lyase (Suy). A high level of sulfite dehydrogenase was also induced during growth with cysteate, and the organism excreted sulfate. A putative regulator, OrfR, was encoded upstream of suyAB on the reverse strand. Downstream of suyAB was suyZ, which was cotranscribed with suyB. -
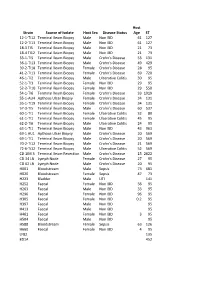
Strain Source of Isolate Host Sex Disease Status Host Age ST
Host Strain Source of Isolate Host Sex Disease Status Age ST 12-1-TI12 Terminal Ileum Biopsy Male Non IBD 61 127 12-2-TI13 Terminal Ileum Biopsy Male Non IBD 61 127 18-3 TI5 Terminal Ileum Biopsy Male Non IBD 21 73 18-4 TI12 Terminal Ileum Biopsy Male Non IBD 21 73 33-1-TI5 Terminal Ileum Biopsy Male Crohn's Disease 53 131 36-1-TI13 Terminal Ileum Biopsy Male Crohn's Disease 49 429 39-2-TI18 Terminal Ileum Biopsy Female Crohn's Disease 28 95 41-2-TI13 Terminal Ileum Biopsy Female Crohn's Disease 69 720 46-1-TI2 Terminal Ileum Biopsy Male Ulcerative Colitis 30 95 52-1-TI3 Terminal Ileum Biopsy Female Non IBD 29 95 52-2-TI10 Terminal Ileum Biopsy Female Non IBD 29 550 54-1-TI6 Terminal Ileum Biopsy Female Crohn's Disease 33 1919 55-1-AU4 Apthous Ulcer Biopsy Female Crohn's Disease 34 131 55-1-TI19 Terminal Ileum Biopsy Female Crohn's Disease 34 131 57-3-TI5 Terminal Ileum Biopsy Male Crohn's Disease 60 537 60-1-TI1 Terminal Ileum Biopsy Female Ulcerative Colitis 32 80 61-1-TI1 Terminal Ileum Biopsy Female Ulcerative Colitis 45 95 62-2-TI6 Terminal Ileum Biopsy Male Ulcerative Colitis 24 95 63-1-TI1 Terminal Ileum Biopsy Male Non IBD 43 963 69-1 AU1 Apthous Ulcer Biopsy Male Crohn's Disease 20 569 69-1-TI1 Terminal Ileum Biopsy Male Crohn's Disease 20 569 70-2-TI12 Terminal Ileum Biopsy Male Crohn's Disease 21 569 72-6-Ti12 Terminal Ileum Biopsy Male Ulcerative Colitis 52 569 CD 1IM 3 Terminal Ileum Resection Male Crohn's Disease 15 2622 CD 34 LN Lymph Node Female Crohn's Disease 27 95 CD 62 LN Lymph Node Male Crohn's Disease 20 95 H001 -

Supplementary Information
Supplementary information (a) (b) Figure S1. Resistant (a) and sensitive (b) gene scores plotted against subsystems involved in cell regulation. The small circles represent the individual hits and the large circles represent the mean of each subsystem. Each individual score signifies the mean of 12 trials – three biological and four technical. The p-value was calculated as a two-tailed t-test and significance was determined using the Benjamini-Hochberg procedure; false discovery rate was selected to be 0.1. Plots constructed using Pathway Tools, Omics Dashboard. Figure S2. Connectivity map displaying the predicted functional associations between the silver-resistant gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S3. Connectivity map displaying the predicted functional associations between the silver-sensitive gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S4. Metabolic overview of the pathways in Escherichia coli. The pathways involved in silver-resistance are coloured according to respective normalized score. Each individual score represents the mean of 12 trials – three biological and four technical. Amino acid – upward pointing triangle, carbohydrate – square, proteins – diamond, purines – vertical ellipse, cofactor – downward pointing triangle, tRNA – tee, and other – circle. -

Kinetic Analysis of Pseudouridine Formation in Trna by Archaeal Cbf5 and Pus10
KINETIC ANALYSIS OF PSEUDOURIDINE FORMATION IN TRNA BY ARCHAEAL CBF5 AND PUS10 RAJASHEKHAR KAMALAMPETA M.Sc. University of Skövde, 2008 A Thesis Submitted to the School of Graduate Studies of the University of Lethbridge in Partial Fulfillment of the Requirements for the Degree DOCTOR OF PHILOSOPHY IN BIOMOLECULAR SCIENCE Department of Chemistry and Biochemistry University of Lethbridge LETHBRIDGE, ALBERTA, CANADA © Rajashekhar Kamalampeta, 2013 Dedicated to My parents for their unconditional love and support & Those passionate teachers for instilling my interest in science iii Abstract Pseudouridines (), the most common modifications in RNA, are formed by stand-alone synthases in all organisms. In addition, archaea and eukaryotes use H/ACA small ribonucleoproteins for pseudouridylation. Cbf5, the catalytic component of these complexes, can also introduce 55 in archaeal tRNAs in a guide RNA-independent manner. Here, kinetic and thermodynamic analyses revealed that both Pyrococcus furiosus Nop10 and Gar1 proteins enhance the catalytic ability of Cbf5 and increase its affinity for tRNA. Pus10, representing a novel synthase family, is the in vivo archaeal tRNA 55 synthase. Characterization of several Pus10 variants demonstrated the importance of the thumb loop for catalysis, a potential role of the THUMP domain in tRNA binding and a new catalytic arginine which may flip the target uridine into Pus10’s active site. The quantitative characterization of the archaeal pseudouridine synthases Cbf5 and Pus10 reported here sheds light on their cellular roles in RNA modification. iv Acknowledgements First and foremost, I would like to express my sincere gratitude to Dr. Ute Kothe, my supervisor, for her invaluable guidance, mentorship, encouragement, and constructive suggestions throughout my Ph.D. -

Supplemental Table 7. Every Significant Association
Supplemental Table 7. Every significant association between an individual covariate and functional group (assigned to the KO level) as determined by CPGLM regression analysis. Variable Unit RelationshipLabel See also CBCL Aggressive Behavior K05914 + CBCL Emotionally Reactive K05914 + CBCL Externalizing Behavior K05914 + K15665 K15658 CBCL Total K05914 + K15660 K16130 KO: E1.13.12.7; photinus-luciferin 4-monooxygenase (ATP-hydrolysing) [EC:1.13.12.7] :: PFAMS: AMP-binding enzyme; CBQ Inhibitory Control K05914 - K12239 K16120 Condensation domain; Methyltransferase domain; Thioesterase domain; AMP-binding enzyme C-terminal domain LEC Family Separation/Social Services K05914 + K16129 K16416 LEC Poverty Related Events K05914 + K16124 LEC Total K05914 + LEC Turmoil K05914 + CBCL Aggressive Behavior K15665 + CBCL Anxious Depressed K15665 + CBCL Emotionally Reactive K15665 + K05914 K15658 CBCL Externalizing Behavior K15665 + K15660 K16130 KO: K15665, ppsB, fenD; fengycin family lipopeptide synthetase B :: PFAMS: Condensation domain; AMP-binding enzyme; CBCL Total K15665 + K12239 K16120 Phosphopantetheine attachment site; AMP-binding enzyme C-terminal domain; Transferase family CBQ Inhibitory Control K15665 - K16129 K16416 LEC Poverty Related Events K15665 + K16124 LEC Total K15665 + LEC Turmoil K15665 + CBCL Aggressive Behavior K11903 + CBCL Anxiety Problems K11903 + CBCL Anxious Depressed K11903 + CBCL Depressive Problems K11903 + LEC Turmoil K11903 + MODS: Type VI secretion system K01220 K01058 CBCL Anxiety Problems K11906 + CBCL Depressive -

Kinetic Analysis of Pseudouridine Formation in Trna by Archaeal Cbf5 and Pus10
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by OPUS: Open Uleth Scholarship - University of Lethbridge Research Repository University of Lethbridge Research Repository OPUS http://opus.uleth.ca Theses Arts and Science, Faculty of 2013 Kinetic analysis of pseudouridine formation in tRNA by archaeal Cbf5 and Pus10 Kamalampeta, Rajashekhar Lethbridge, Alta. : University of Lethbridge, Dept. of Chemistry and Biochemistry http://hdl.handle.net/10133/3464 Downloaded from University of Lethbridge Research Repository, OPUS KINETIC ANALYSIS OF PSEUDOURIDINE FORMATION IN TRNA BY ARCHAEAL CBF5 AND PUS10 RAJASHEKHAR KAMALAMPETA M.Sc. University of Skövde, 2008 A Thesis Submitted to the School of Graduate Studies of the University of Lethbridge in Partial Fulfillment of the Requirements for the Degree DOCTOR OF PHILOSOPHY IN BIOMOLECULAR SCIENCE Department of Chemistry and Biochemistry University of Lethbridge LETHBRIDGE, ALBERTA, CANADA © Rajashekhar Kamalampeta, 2013 Dedicated to My parents for their unconditional love and support & Those passionate teachers for instilling my interest in science iii Abstract Pseudouridines (), the most common modifications in RNA, are formed by stand-alone synthases in all organisms. In addition, archaea and eukaryotes use H/ACA small ribonucleoproteins for pseudouridylation. Cbf5, the catalytic component of these complexes, can also introduce 55 in archaeal tRNAs in a guide RNA-independent manner. Here, kinetic and thermodynamic analyses revealed that both Pyrococcus furiosus Nop10 and Gar1 proteins enhance the catalytic ability of Cbf5 and increase its affinity for tRNA. Pus10, representing a novel synthase family, is the in vivo archaeal tRNA 55 synthase. Characterization of several Pus10 variants demonstrated the importance of the thumb loop for catalysis, a potential role of the THUMP domain in tRNA binding and a new catalytic arginine which may flip the target uridine into Pus10’s active site. -

Novel Non-Phosphorylative Pathway of Pentose Metabolism from Bacteria
www.nature.com/scientificreports OPEN Novel non-phosphorylative pathway of pentose metabolism from bacteria Received: 22 March 2018 Seiya Watanabe1,2,3, Fumiyasu Fukumori4, Hisashi Nishiwaki1,2, Yasuhiro Sakurai5, Accepted: 30 September 2018 Kunihiko Tajima5 & Yasuo Watanabe1,2 Published: xx xx xxxx Pentoses, including D-xylose, L-arabinose, and D-arabinose, are generally phosphorylated to D-xylulose 5-phosphate in bacteria and fungi. However, in non-phosphorylative pathways analogous to the Entner-Dodorof pathway in bacteria and archaea, such pentoses can be converted to pyruvate and glycolaldehyde (Route I) or α-ketoglutarate (Route II) via a 2-keto-3-deoxypentonate (KDP) intermediate. Putative gene clusters related to these metabolic pathways were identifed on the genome of Herbaspirillum huttiense IAM 15032 using a bioinformatic analysis. The biochemical characterization of C785_RS13685, one of the components encoded to D-arabinonate dehydratase, difered from the known acid-sugar dehydratases. The biochemical characterization of the remaining components and a genetic expression analysis revealed that D- and L-KDP were converted not only to α-ketoglutarate, but also pyruvate and glycolate through the participation of dehydrogenase and hydrolase (Route III). Further analyses revealed that the Route II pathway of D-arabinose metabolism was not evolutionally related to the analogous pathway from archaea. Te breakdown of D-glucose is central for energy and biosynthetic metabolism throughout all domains of life. Te most common glycolytic routes in bacteria are the Embden-Meyerhof-Parnas, the Entner-Doudorof (ED), and the oxidative pentose phosphate pathways. Te distinguishing diference between the two former glyco- lytic pathways lies in the nature of the 6-carbon metabolic intermediates that serve as substrates for aldol cleav- age. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

The HDHD1 Gene, Often Deleted in X-Linked Ichthyosis, Encodes a Pseudouridine-5’-Phosphatase Alice Preumont, Rim Rzem, Didier Vertommen, Emile Van Schaftingen
The HDHD1 gene, often deleted in X-linked ichthyosis, encodes a pseudouridine-5’-phosphatase Alice Preumont, Rim Rzem, Didier Vertommen, Emile van Schaftingen To cite this version: Alice Preumont, Rim Rzem, Didier Vertommen, Emile van Schaftingen. The HDHD1 gene, often deleted in X-linked ichthyosis, encodes a pseudouridine-5’-phosphatase. Biochemical Journal, Portland Press, 2010, 431 (2), pp.237-244. 10.1042/BJ20100174. hal-00521555 HAL Id: hal-00521555 https://hal.archives-ouvertes.fr/hal-00521555 Submitted on 28 Sep 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Biochemical Journal Immediate Publication. Published on 20 Aug 2010 as manuscript BJ20100174 The HDHD1 gene, often deleted in X-linked ichthyosis, encodes a pseudouridine-5’- phosphatase Alice Preumont, Rim Rzem, Didier Vertommen, Emile Van Schaftingen From the de Duve Institute, Université Catholique de Louvain, B-1200 Brussels, Belgium Address correspondence to: Emile Van Schaftingen, de Duve Institute, Université catholique de Louvain, Avenue Hippocrate 75, B-1200 Brussels, Belgium. Tel.: 3227647564; Fax: 3227647598; E-mail: [email protected] The abbreviations used are: 5’-PsiMP, pseudouridine 5’-phosphate; 5’-PsiMPase, pseudouridine-5’-phosphatase; DTT, dithiothreitol; FMNPase, FMN phosphatase ; LB, Luria- Bertani. -

POLSKIE TOWARZYSTWO BIOCHEMICZNE Postępy Biochemii
POLSKIE TOWARZYSTWO BIOCHEMICZNE Postępy Biochemii http://rcin.org.pl WSKAZÓWKI DLA AUTORÓW Kwartalnik „Postępy Biochemii” publikuje artykuły monograficzne omawiające wąskie tematy, oraz artykuły przeglądowe referujące szersze zagadnienia z biochemii i nauk pokrewnych. Artykuły pierwszego typu winny w sposób syntetyczny omawiać wybrany temat na podstawie możliwie pełnego piśmiennictwa z kilku ostatnich lat, a artykuły drugiego typu na podstawie piśmiennictwa z ostatnich dwu lat. Objętość takich artykułów nie powinna przekraczać 25 stron maszynopisu (nie licząc ilustracji i piśmiennictwa). Kwartalnik publikuje także artykuły typu minireviews, do 10 stron maszynopisu, z dziedziny zainteresowań autora, opracowane na podstawie najnow szego piśmiennictwa, wystarczającego dla zilustrowania problemu. Ponadto kwartalnik publikuje krótkie noty, do 5 stron maszynopisu, informujące o nowych, interesujących osiągnięciach biochemii i nauk pokrewnych, oraz noty przybliżające historię badań w zakresie różnych dziedzin biochemii. Przekazanie artykułu do Redakcji jest równoznaczne z oświadczeniem, że nadesłana praca nie była i nie będzie publikowana w innym czasopiśmie, jeżeli zostanie ogłoszona w „Postępach Biochemii”. Autorzy artykułu odpowiadają za prawidłowość i ścisłość podanych informacji. Autorów obowiązuje korekta autorska. Koszty zmian tekstu w korekcie (poza poprawieniem błędów drukarskich) ponoszą autorzy. Artykuły honoruje się według obowiązujących stawek. Autorzy otrzymują bezpłatnie 25 odbitek swego artykułu; zamówienia na dodatkowe odbitki (płatne) należy zgłosić pisemnie odsyłając pracę po korekcie autorskiej. Redakcja prosi autorów o przestrzeganie następujących wskazówek: Forma maszynopisu: maszynopis pracy i wszelkie załączniki należy nadsyłać w dwu egzem plarzach. Maszynopis powinien być napisany jednostronnie, z podwójną interlinią, z marginesem ok. 4 cm po lewej i ok. 1 cm po prawej stronie; nie może zawierać więcej niż 60 znaków w jednym wierszu nie więcej niż 30 wierszy na stronie zgodnie z Normą Polską. -

Regulatory Role of Plar (Yiaj) for Plant Utilization in Escherichia Coli K-12
www.nature.com/scientificreports Corrected: Author Correction OPEN Regulatory Role of PlaR (YiaJ) for Plant Utilization in Escherichia coli K-12 Tomohiro Shimada 1,2*, Yui Yokoyama3, Takumi Anzai1, Kaneyoshi Yamamoto3 & Akira Ishihama2,3* Outside a warm-blooded animal host, the enterobacterium Escherichia coli K-12 is also able to grow and survive in stressful nature. The major organic substance in nature is plant, but the genetic system of E. coli how to utilize plant-derived materials as nutrients is poorly understood. Here we describe the set of regulatory targets for uncharacterized IclR-family transcription factor YiaJ on the E. coli genome, using gSELEX screening system. Among a total of 18 high-afnity binding targets of YiaJ, the major regulatory target was identifed to be the yiaLMNOPQRS operon for utilization of ascorbate from fruits and galacturonate from plant pectin. The targets of YiaJ also include the genes involved in the utilization for other plant-derived materials as nutrients such as fructose, sorbitol, glycerol and fructoselysine. Detailed in vitro and in vivo analyses suggest that L-ascorbate and α-D-galacturonate are the efector ligands for regulation of YiaJ function. These fndings altogether indicate that YiaJ plays a major regulatory role in expression of a set of the genes for the utilization of plant-derived materials as nutrients for survival. PlaR was also suggested to play protecting roles of E. coli under stressful environments in nature, including the formation of bioflm. We then propose renaming YiaJ to PlaR (regulator of plant utilization). Bacteria constantly monitor environmental conditions, and respond for adaptation and survival by modulating the expression pattern of their genomes.