Screening for Differentially Expressed Genes Between Left‑And Right‑Sided
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Expression Profiling of KLF4
Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in wild type MEFs. 98 Am J Cancer Res 2011;1(1):85-97 Table S1: Functional Annotation Clustering of Genes Up-Regulated in Klf4 -Null MEFs ILLUMINA_ID Gene Symbol Gene Name (Description) P -value Fold-Change Cell Cycle 8.00E-03 ILMN_1217331 Mcm6 MINICHROMOSOME MAINTENANCE DEFICIENT 6 40.36 ILMN_2723931 E2f6 E2F TRANSCRIPTION FACTOR 6 26.8 ILMN_2724570 Mapk12 MITOGEN-ACTIVATED PROTEIN KINASE 12 22.19 ILMN_1218470 Cdk2 CYCLIN-DEPENDENT KINASE 2 9.32 ILMN_1234909 Tipin TIMELESS INTERACTING PROTEIN 5.3 ILMN_1212692 Mapk13 SAPK/ERK/KINASE 4 4.96 ILMN_2666690 Cul7 CULLIN 7 2.23 ILMN_2681776 Mapk6 MITOGEN ACTIVATED PROTEIN KINASE 4 2.11 ILMN_2652909 Ddit3 DNA-DAMAGE INDUCIBLE TRANSCRIPT 3 2.07 ILMN_2742152 Gadd45a GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE 45 ALPHA 1.92 ILMN_1212787 Pttg1 PITUITARY TUMOR-TRANSFORMING 1 1.8 ILMN_1216721 Cdk5 CYCLIN-DEPENDENT KINASE 5 1.78 ILMN_1227009 Gas2l1 GROWTH ARREST-SPECIFIC 2 LIKE 1 1.74 ILMN_2663009 Rassf5 RAS ASSOCIATION (RALGDS/AF-6) DOMAIN FAMILY 5 1.64 ILMN_1220454 Anapc13 ANAPHASE PROMOTING COMPLEX SUBUNIT 13 1.61 ILMN_1216213 Incenp INNER CENTROMERE PROTEIN 1.56 ILMN_1256301 Rcc2 REGULATOR OF CHROMOSOME CONDENSATION 2 1.53 Extracellular Matrix 5.80E-06 ILMN_2735184 Col18a1 PROCOLLAGEN, TYPE XVIII, ALPHA 1 51.5 ILMN_1223997 Crtap CARTILAGE ASSOCIATED PROTEIN 32.74 ILMN_2753809 Mmp3 MATRIX METALLOPEPTIDASE -

Increased Expression of Unmethylated CDKN2D by 5-Aza-2'-Deoxycytidine in Human Lung Cancer Cells
Oncogene (2001) 20, 7787 ± 7796 ã 2001 Nature Publishing Group All rights reserved 0950 ± 9232/01 $15.00 www.nature.com/onc Increased expression of unmethylated CDKN2D by 5-aza-2'-deoxycytidine in human lung cancer cells Wei-Guo Zhu1, Zunyan Dai2,3, Haiming Ding1, Kanur Srinivasan1, Julia Hall3, Wenrui Duan1, Miguel A Villalona-Calero1, Christoph Plass3 and Gregory A Otterson*,1 1Division of Hematology/Oncology, Department of Internal Medicine, The Ohio State University-Comprehensive Cancer Center, Columbus, Ohio, OH 43210, USA; 2Department of Pathology, The Ohio State University-Comprehensive Cancer Center, Columbus, Ohio, OH 43210, USA; 3Division of Human Cancer Genetics, Department of Molecular Virology, Immunology and Medical Genetics, The Ohio State University-Comprehensive Cancer Center, Columbus, Ohio, OH 43210, USA DNA hypermethylation of CpG islands in the promoter Introduction region of genes is associated with transcriptional silencing. Treatment with hypo-methylating agents can Methylation of cytosine residues in CpG sequences is a lead to expression of these silenced genes. However, DNA modi®cation that plays a role in normal whether inhibition of DNA methylation in¯uences the mammalian development (Costello and Plass, 2001; expression of unmethylated genes has not been exten- Li et al., 1992), imprinting (Li et al., 1993) and X sively studied. We analysed the methylation status of chromosome inactivation (Pfeifer et al., 1990). To date, CDKN2A and CDKN2D in human lung cancer cell lines four mammalian DNA methyltransferases (DNMT) and demonstrated that the CDKN2A CpG island is have been identi®ed (Bird and Wole, 1999). Disrup- methylated, whereas CDKN2D is unmethylated. Treat- tion of the balance in methylated DNA is a common ment of cells with 5-aza-2'-deoxycytidine (5-Aza-CdR), alteration in cancer (Costello et al., 2000; Costello and an inhibitor of DNA methyltransferase 1, induced a dose Plass, 2001; Issa et al., 1993; Robertson et al., 1999). -

Deletions of CDKN2C in Multiple Myeloma: Biological and Clinical Implications Paola E
Human Cancer Biology Deletions of CDKN2C in Multiple Myeloma: Biological and Clinical Implications Paola E. Leone,1Brian A. Walker,1Matthew W. Jenner,1Laura Chiecchio,2 GianPaolo Dagrada,2 Rebecca K.M. Protheroe,2 David C. Johnson,1Nicholas J. Dickens,1Jose Luis Brito,1Monica Else,1 David Gonzalez,1Fiona M. Ross,2 Selina Chen-Kiang,3 Faith E. Davies,1and Gareth J. Morgan1 Abstract Purpose: Deletions of chromosome 1have been described in 7% to 40% of cases of myeloma with inconsistent clinical consequences. CDKN2C at 1p32.3 has been identified in myeloma cell lines as the potential target of the deletion.We tested the clinical impact of 1p deletion and used high-resolution techniques to define the role of CDKN2C in primary patient material. Experimental Design: We analyzed 515 cases of monoclonal gammopathy of undetermined significance (MGUS), smoldering multiple myeloma (SMM), and newly diagnosed multiple mye- loma using fluorescence in situ hybridization (FISH) for deletions of CDKN2C.In78myeloma cases, we carried out Affymetrix single nucleotide polymorphism mapping and U133 Plus 2.0 ex- pression arrays. In addition, we did mutation, methylation, andWestern blotting analysis. Results: By FISH we identified deletion of 1p32.3 (CDKN2C)in3of66MGUS(4.5%),4of39 SMM (10.3%), and 55 of 369 multiple myeloma cases (15%).We examined the impact of copy number change at CDKN2C on overall survival (OS), and found that the cases with either hemi- zygous or homozygous deletion of CDKN2C had a worse OS compared with cases that were intact at this region (22 months versus 38 months; P = 0.003). Using gene mapping we identified three homozygous deletions at 1p32.3, containing CDKN2C,allofwhichlackedexpressionof CDKN2C. -

P63 Modulates the Expression of the WDFY2 Gene Which Is Implicated in Cancer Regulation and Limb Development
Bioscience Reports (2019) 39 BSR20192114 https://doi.org/10.1042/BSR20192114 Research Article P63 modulates the expression of the WDFY2 gene which is implicated in cancer regulation and limb development 1 2 1,* 1 2 3 Paola Monti , Yari Ciribilli , Giorgia Foggetti , Paola Menichini , Alessandra Bisio , Serena Cappato , Downloaded from https://portlandpress.com/bioscirep/article-pdf/39/12/BSR20192114/862944/bsr-2019-2114.pdf by guest on 07 February 2020 Alberto Inga2, Maria Teresa Divizia4, Margherita Lerone4, Renata Bocciardi3,4 and Gilberto Fronza1 1Mutagenesis and Cancer Prevention Unit, IRCCS Ospedale Policlinico San Martino, Largo R. Benzi, 10, Genoa 16132, Italy; 2Department of Cellular, Computational and Integrative Biology (CIBio), University of Trento, Via Sommarive 9, Povo (TN) 38123, Italy; 3Department of Neurosciences, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health (DiNOGMI), University of Genoa, Largo P. Daneo, 3, Genoa 16132, Italy; 4Medical Genetics Unit, IRCCS Istituto Giannina Gaslini, Via G. Gaslini, 5, Genoa 16147, Italy Correspondence: Paola Monti ([email protected]) or Gilberto Fronza ([email protected]) TP63 is a member of the TP53 gene family, sharing a common gene structure that produces two groups of mRNAs’ encoding proteins with different N-terminal regions (N and TA iso- forms); both transcripts are also subjected to alternative splicing mechanisms at C-terminus, generating a variety of isoforms. p63 is a master regulator of epidermal development and homoeostasis as well as an important player in tumorigenesis and cancer progression with both oncogenic and tumour suppressive roles. A number of studies have aimed at the identi- fication of p63 target genes, allowing the dissection of the molecular pathways orchestrated by the different isoforms. -

(CDKN2D) (NM 001800) Human Recombinant Protein Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP314065 p19 INK4d (CDKN2D) (NM_001800) Human Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Recombinant protein of human cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (CDKN2D), transcript variant 1 Species: Human Expression Host: HEK293T Tag: C-Myc/DDK Predicted MW: 17.5 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol Preparation: Recombinant protein was captured through anti-DDK affinity column followed by conventional chromatography steps. Storage: Store at -80°C. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_001791 Locus ID: 1032 UniProt ID: P55273, A0A024R796 RefSeq Size: 1416 Cytogenetics: 19p13.2 RefSeq ORF: 498 Synonyms: INK4D; p19; p19-INK4D This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 p19 INK4d (CDKN2D) (NM_001800) Human Recombinant Protein – TP314065 Summary: The protein encoded by this gene is a member of the INK4 family of cyclin-dependent kinase inhibitors. This protein has been shown to form a stable complex with CDK4 or CDK6, and prevent the activation of the CDK kinases, thus function as a cell growth regulator that controls cell cycle G1 progression. -

Loss of the Nuclear Wnt Pathway Effector TCF7L2 Promotes Migration and Invasion of Human Colorectal Cancer Cells
Oncogene (2020) 39:3893–3909 https://doi.org/10.1038/s41388-020-1259-7 ARTICLE Loss of the nuclear Wnt pathway effector TCF7L2 promotes migration and invasion of human colorectal cancer cells 1,2 1 3 4,5,6 2,5 Janna Wenzel ● Katja Rose ● Elham Bavafaye Haghighi ● Constanze Lamprecht ● Gilles Rauen ● 1 7 3,8,9 1,2,5 Vivien Freihen ● Rebecca Kesselring ● Melanie Boerries ● Andreas Hecht Received: 27 September 2019 / Revised: 3 March 2020 / Accepted: 4 March 2020 / Published online: 20 March 2020 © The Author(s) 2020. This article is published with open access Abstract The transcription factor TCF7L2 is indispensable for intestinal tissue homeostasis where it transmits mitogenic Wnt/ β-Catenin signals in stem and progenitor cells, from which intestinal tumors arise. Yet, TCF7L2 belongs to the most frequently mutated genes in colorectal cancer (CRC), and tumor-suppressive functions of TCF7L2 were proposed. This apparent paradox warrants to clarify the role of TCF7L2 in colorectal carcinogenesis. Here, we investigated TCF7L2 dependence/independence of CRC cells and the cellular and molecular consequences of TCF7L2 loss-of-function. By genome editing we achieved complete TCF7L2 inactivation in several CRC cell lines without loss of viability, showing that fi 1234567890();,: 1234567890();,: CRC cells have widely lost the strict requirement for TCF7L2. TCF7L2 de ciency impaired G1/S progression, reminiscent of the physiological role of TCF7L2. In addition, TCF7L2-negative cells exhibited morphological changes, enhanced migration, invasion, and collagen adhesion, albeit the severity of the phenotypic alterations manifested in a cell-line-specific fashion. To provide a molecular framework for the observed cellular changes, we performed global transcriptome profiling and identified gene-regulatory networks in which TCF7L2 positively regulates the proto-oncogene MYC, while repressing the cell cycle inhibitors CDKN2C/CDKN2D. -

Inhibition of CDK4/6 As Therapeutic Approach for Ovarian Cancer Patients: Current Evidences and Future Perspectives
cancers Review Inhibition of CDK4/6 as Therapeutic Approach for Ovarian Cancer Patients: Current Evidences and Future Perspectives Alessandra Dall’Acqua 1,†, Michele Bartoletti 2,3,† , Nastaran Masoudi-Khoram 1,‡, Roberto Sorio 2, Fabio Puglisi 2,3 , Barbara Belletti 1 and Gustavo Baldassarre 1,* 1 Molecular Oncology Unit, Centro di Riferimento Oncologico di Aviano (CRO), IRCCS, National Cancer Institute, 33081 Aviano, Italy; [email protected] (A.D.); [email protected] (N.M.-K.); [email protected] (B.B.) 2 Medical Oncology and Cancer Prevention Molecular Oncology Unit, Centro di Riferimento Oncologico di Aviano (CRO), IRCCS, National Cancer Institute, 33081 Aviano, Italy; [email protected] (M.B.); [email protected] (R.S.); [email protected] (F.P.) 3 Department of Medicine (DAME), University of Udine, 33100 Udine, Italy * Correspondence: [email protected]; Tel.: +39-0434-659779; Fax: +39-0434-659429 † These authors equally contributed to the work. ‡ Present address: Department of Biophysics, Faculty of Biological Sciences, Tarbiat Modares University, Tehran, Iran. Simple Summary: Altered regulation of the cell cycle is a hallmark of cancer. The recent clinical success of the inhibitors of CDK4 and CDK6 has convincingly demonstrated that targeting cell cycle components may represent an effective anti-cancer strategy, at least in some cancer types. However, possible applications of CDK4/6 inhibitors in patients with ovarian cancer is still under evaluation. Citation: Dall’Acqua, A.; Bartoletti, M.; Masoudi-Khoram, N.; Sorio, R.; Here, we describe the possible biological role of CDK4 and CDK6 complexes in ovarian cancer and Puglisi, F.; Belletti, B.; Baldassarre, G. provide the rationale for the use of CDK4/6 inhibitors in this pathology, alone or in combination Inhibition of CDK4/6 as Therapeutic with other drugs. -

Loss of Expression of Cyclin D2 by Aberrant DNA Methylation: a Potential Biomarker in Vietnamese Breast Cancer Patients
DOI:http://dx.doi.org/10.7314/APJCP.2015.16.6.2209 Loss of Expression of Cyclin D2 by Aberrant DNA Methylation: a Potential Biomarker in Vietnamese Breast Cancer Patients RESEARCH ARTICLE Loss of Expression of Cyclin D2 by Aberrant DNA Methylation: a Potential Biomarker in Vietnamese Breast Cancer Patients Phuong Kim Truong1, Thuan Duc Lao1, Thao Phuong Thi Doan2, Thuy Ai Huyen Le1* Abstract DNA methylation of tumor suppressor gene promoters is the most frequent phenomenon leading to inactivation of function, consequently driving malignant cell transformation. Cyclin D2 is implicated in tumor suppression. In our study, we carried out the MSP assay to evaluation the methylation status at CpG islands in the cyclin D2 promoter in breast cancer cases from the Vietnamese population. The results showed that the frequency of methylation reached 62.1% (59 of 95 breast cancer tumors), but was low in non-cancer specimens at 10% (2 of 20 non-cancer specimens). Additionally, with an RR (relative risk) and OR (odd ratios) of 6.21 and 14.8, DNA hypermethylation of cyclin D2 increased the possibility of malignant transformation. Our results confirmed the cyclin D2 hypermethylation could be used as the potential biomarker which could be applied in prognosis and early diagnosis of Vietnamese breast cancer patients. Keywords: Cyclin D2 - methylation - MSP - breast cancer - Vietnamese population Asian Pac J Cancer Prev, 16 (6), 2209-2213 Introduction is the covalent addition of the methyl group to Cytosine residues of the CpG dinucleotide in DNA, which plays Cyclin D2, a protein belonging to the highly conserved an important role in driving tumorgenesis, including cyclin family, is characterized as the regulator of the cell breast carcinoma (Pongtheerat et al., 2011; Ramezani et cycle at the point of transition from G1 to S phase during al., 2012; Phuong et al., 2014). -

The Role of Cdks and Cdkis in Murine Development
International Journal of Molecular Sciences Review The Role of CDKs and CDKIs in Murine Development Grace Jean Campbell , Emma Langdale Hands and Mathew Van de Pette * Epigenetic Mechanisms of Toxicology Lab, MRC Toxicology Unit, Cambridge University, Cambridge CB2 1QR, UK; [email protected] (G.J.C.); [email protected] (E.L.H.) * Correspondence: [email protected] Received: 8 July 2020; Accepted: 26 July 2020; Published: 28 July 2020 Abstract: Cyclin-dependent kinases (CDKs) and their inhibitors (CDKIs) play pivotal roles in the regulation of the cell cycle. As a result of these functions, it may be extrapolated that they are essential for appropriate embryonic development. The twenty known mouse CDKs and eight CDKIs have been studied to varying degrees in the developing mouse, but only a handful of CDKs and a single CDKI have been shown to be absolutely required for murine embryonic development. What has become apparent, as more studies have shone light on these family members, is that in addition to their primary functional role in regulating the cell cycle, many of these genes are also controlling specific cell fates by directing differentiation in various tissues. Here we review the extensive mouse models that have been generated to study the functions of CDKs and CDKIs, and discuss their varying roles in murine embryonic development, with a particular focus on the brain, pancreas and fertility. Keywords: cyclin-dependent kinase; CDK inhibitors; mouse; development; knock-out models 1. Introduction Cyclin-dependent kinases (CDKs) are proteins that, by definition, require the binding of partner cyclin proteins in order to phosphorylate a series of target proteins. -

Human CDKN2D / P19ink4d Protein (GST Tag)
Human CDKN2D / p19ink4d Protein (GST Tag) Catalog Number: 12558-H09E General Information SDS-PAGE: Gene Name Synonym: INK4D; p19; p19-INK4D Protein Construction: A DNA sequence encoding the human CDKN2D (P55273) (Met 10Leu 166) was fused with the GST tag at the N-terminus. Source: Human Expression Host: E. coli QC Testing Purity: > 90 % as determined by SDS-PAGE Bio Activity: Protein Description Immobilized human GST-CDKN2D at 10 μg/ml (100 μl/well) can bind biotinylated human GST-CDK4 (Cat:10732-H09B), The EC50 of Cyclin-dependent kinase inhibitor 2D(also known as CDKN2D or p19ink4d), biotinylated human GST-CDK4 (Cat:10732-H09B) is 0.52-1.2 μg/ml. a member of the INK4 family of cyclin-dependent kinase (CDK) inhibitors, negatively regulates the cyclin D-CDK4/6 complexes, which promote G1/S Endotoxin: transition by phosphorylating the retinoblastoma tumor-suppressor gene product. It is clearly shown that DNA repair is the main target of p19ink4d Please contact us for more information. effect and that diminished apoptosis is a downstream event. Experiments has uncovered a role of p19INK4d as a regulator of DNA-damage-induced Stability: apoptosis and suggest that it protects cells from undergoing apoptosis by Samples are stable for up to twelve months from date of receipt at -70 ℃ allowing a more efficient DNA repair. It has been demonstrated that p19INK4d expression enhances cell survival under genotoxic conditions. Predicted N terminal: Met Previous work has shown that inactivation of the cyclin-dependent kinase inhibitor (CKI) p19(Ink4d) leads to progressive hearing loss attributable to Molecular Mass: inappropriate DNA replication and subsequent apoptosis of hair cells. -

MEN4 and CDKN1B Mutations 24:10 T195–T208 Thematic Review
2410 R Alrezk et al. MEN4 and CDKN1B mutations 24:10 T195–T208 Thematic Review MEN4 and CDKN1B mutations: the latest of the MEN syndromes Rami Alrezk1, Fady Hannah-Shmouni2 and Constantine A Stratakis2 1 The National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Correspondence Maryland, USA should be addressed 2 Section on Endocrinology & Genetics, the Eunice Kennedy Shriver National Institute of Child Health and Human to C A Stratakis Development, NIH, Bethesda, Maryland, USA Email [email protected] Abstract Multiple endocrine neoplasia (MEN) refers to a group of autosomal dominant disorders Key Words with generally high penetrance that lead to the development of a wide spectrum of f multiple endocrine endocrine and non-endocrine manifestations. The most frequent among these conditions neoplasia is MEN type 1 (MEN1), which is caused by germline heterozygous loss-of-function f MEN4 mutations in the tumor suppressor gene MEN1. MEN1 is characterized by primary f MEN1 hyperparathyroidism (PHPT) and functional or nonfunctional pancreatic neuroendocrine f neuroendocrine tumors tumors and pituitary adenomas. Approximately 10% of patients with familial or sporadic f CDKN1B MEN1-like phenotype do not have MEN1 mutations or deletions. A novel MEN syndrome f p27 was discovered, initially in rats (MENX), and later in humans (MEN4), which is caused by germline mutations in the putative tumor suppressor CDKN1B. The most common phenotype of the 19 established cases of MEN4 that have been described to date is PHPT Endocrine-Related Cancer Endocrine-Related followed by pituitary adenomas. Recently, somatic or germline mutations in CDKN1B were also identified in patients with sporadic PHPT, small intestinal neuroendocrine tumors, lymphoma and breast cancer, demonstrating a novel role for CDKN1B as a tumor susceptibility gene for other neoplasms. -
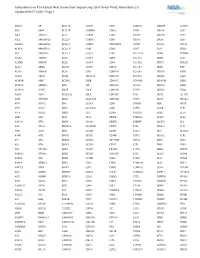
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1 ABCC3 AR BCL11A CANT1 CDK1 CMKLR1 DAB2IP DUSP9 ABI1 ARAF BCL11B CAPRIN1 CDK12 CNBP DACH1 E2F1 ABL1 ARFRP1 BCL2 CAPZB CDK2 CNOT2 DACH2 E2F3 ABL2 ARHGAP20 BCL2A1 CARD11 CDK4 CNTN1 DAXX EBF1 ABLIM1 ARHGAP26 BCL2L1 CARM1 CDK5RAP2 CNTRL DCLK2 ECT2L ACACA ARHGEF12 BCL2L11 CARS CDK6 COG5 DCN EDIL3 ACE ARHGEF7 BCL2L2 CASC5 CDK7 COL11A1 DDB1 EDNRB ACER1 ARID1A BCL3 CASP3 CDK8 COL1A1 DDB2 EED ACSBG1 ARID1B BCL6 CASP7 CDK9 COL1A2 DDIT3 EEFSEC ACSL3 ARID2 BCL7A CASP8 CDKL5 COL3A1 DDR2 EGF ACSL6 ARID5B BCL9 CAV1 CDKN1A COL6A3 DDX10 EGFR ACVR1 ARIH2 BCOR CBFA2T3 CDKN1B COL9A3 DDX20 EGR1 ACVR1B ARNT BCORL1 CBFB CDKN1C COMMD1 DDX39B EGR2 ACVR1C ARRDC4 BCR CBL CDKN2A COX6C DDX3X EGR3 ACVR2A ASMTL BDNF CBLB CDKN2B CPNE1 DDX41 EGR4 ADD3 ASPH BHLHE22 CBLC CDKN2C CPS1 DDX5 EIF1AX ADM ASPSCR1 BICC1 CCDC28A CDKN2D CPSF6 DDX6 EIF4A2 AFF1 ASTN2 BIN1 CCDC6 CDX1 CRADD DEK EIF4E AFF3 ASXL1 BIRC3 CCDC88C CDX2 CREB1 DGKB ELF3 AFF4 ASXL2 BIRC6 CCK CEBPA CREB3L1 DGKI ELF4 AGR3 ATF1 BLM CCL2 CEBPB CREB3L2 DGKZ ELK4 AHCYL1 ATF3 BMP4 CCNA2 CEBPD CREBBP DICER1 ELL AHI1 ATG13 BMPR1A CCNB1IP1 CEBPE CRKL DIRAS3 ELN AHR ATG5 BRAF CCNB3 CENPF CRLF2 DIS3 ELOVL2 AHRR ATIC BRCA1 CCND1 CENPU CRTC1 DIS3L2 ELP2 AIP ATL1 BRCA2 CCND2 CEP170B CRTC3 DKK1 EML1 AK2 ATM BRCC3 CCND3 CEP57 CSF1 DKK2 EML4 AK5 ATP1B4 BRD1 CCNE1 CEP85L CSF1R DKK4 ENPP2 AKAP12 ATP8A2 BRD3 CCNG1 CHCHD7 CSF3 DLEC1 EP300 AKAP6 ATR BRD4 CCT6B CHD2 CSF3R DLL1 EP400 AKAP9 ATRNL1 BRIP1 CD19 CHD4 CSNK1A1 DLL3