For Personal Use. Only Reproduce with Permission from the Lancet Publishing Group
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

Induction of Erythropoietin Increases the Cell Proliferation Rate in a Hypoxia‑Inducible Factor‑1‑Dependent and ‑Independent Manner in Renal Cell Carcinoma Cell Lines
ONCOLOGY LETTERS 5: 1765-1770, 2013 Induction of erythropoietin increases the cell proliferation rate in a hypoxia‑inducible factor‑1‑dependent and ‑independent manner in renal cell carcinoma cell lines YUTAKA FUJISUE1, TAKATOSHI NAKAGAWA2, KIYOSHI TAKAHARA1, TERUO INAMOTO1, SATOSHI KIYAMA1, HARUHITO AZUMA1 and MICHIO ASAHI2 Departments of 1Urology and 2Pharmacology, Faculty of Medicine, Osaka Medical College, Takatsuki, Osaka 569-8686, Japan Received November 28, 2012; Accepted February 25, 2013 DOI: 10.3892/ol.2013.1283 Abstract. Erythropoietin (Epo) is a potent inducer of erythro- Introduction poiesis that is mainly produced in the kidney. Epo is expressed not only in the normal kidney, but also in renal cell carcinomas Erythropoietin (Epo) is a 30-kDa glycoprotein that functions (RCCs). The aim of the present study was to gain insights as an important cytokine in erythrocytes. Epo is usually into the roles of Epo and its receptor (EpoR) in RCC cells. produced by stromal cells of the adult kidney cortex or fetal The study used two RCC cell lines, Caki-1 and SKRC44, in liver and then released into the blood, with its production which Epo and EpoR are known to be highly expressed. The initially induced by hypoxia or hypotension (1-5). In the bone proliferation rate and expression level of hypoxia-inducible marrow, Epo binds to the erythropoietin receptor (EpoR) factor-1α (HIF-1α) were measured prior to and following expressed in erythroid progenitor cells or undifferentiated Epo treatment and under normoxic and hypoxic conditions. erythroblasts, which induces signal transduction mechanisms To examine whether HIF-1α or Epo were involved in cellular that protect the undifferentiated erythrocytes from apoptosis proliferation during hypoxia, these proteins were knocked and promote their proliferation and differentiation. -

Receptor-Arrestin Interactions: the GPCR Perspective
biomolecules Review Receptor-Arrestin Interactions: The GPCR Perspective Mohammad Seyedabadi 1,2 , Mehdi Gharghabi 3, Eugenia V. Gurevich 4 and Vsevolod V. Gurevich 4,* 1 Department of Toxicology & Pharmacology, Faculty of Pharmacy, Mazandaran University of Medical Sciences, Sari 48471-93698, Iran; [email protected] 2 Pharmaceutical Sciences Research Center, Faculty of Pharmacy, Mazandaran University of Medical Sciences, Sari 48167-75952, Iran 3 Department of Cancer Biology and Genetics, The Ohio State University Wexner Medical Center, Columbus, OH 43210, USA; [email protected] 4 Department of Pharmacology, Vanderbilt University, Nashville, TN 37232, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-615-322-7070; Fax: +1-615-343-6532 Abstract: Arrestins are a small family of four proteins in most vertebrates that bind hundreds of different G protein-coupled receptors (GPCRs). Arrestin binding to a GPCR has at least three functions: precluding further receptor coupling to G proteins, facilitating receptor internalization, and initiating distinct arrestin-mediated signaling. The molecular mechanism of arrestin–GPCR interactions has been extensively studied and discussed from the “arrestin perspective”, focusing on the roles of arrestin elements in receptor binding. Here, we discuss this phenomenon from the “receptor perspective”, focusing on the receptor elements involved in arrestin binding and empha- sizing existing gaps in our knowledge that need to be filled. It is vitally important to understand the role of receptor elements in arrestin activation and how the interaction of each of these elements with arrestin contributes to the latter’s transition to the high-affinity binding state. A more precise knowledge of the molecular mechanisms of arrestin activation is needed to enable the construction of arrestin mutants with desired functional characteristics. -

Molecular Pathways Associated with the Nutritional Programming of Plant-Based Diet Acceptance in Rainbow Trout Following an Early Feeding Exposure Mukundh N
Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, Aurélie Le Cam, Françoise Médale, Sadasivam J. Kaushik, Inge Geurden To cite this version: Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, et al.. Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure. BMC Genomics, BioMed Central, 2016, 17 (1), pp.1-20. 10.1186/s12864-016-2804-1. hal-01346912 HAL Id: hal-01346912 https://hal.archives-ouvertes.fr/hal-01346912 Submitted on 19 Jul 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Balasubramanian et al. BMC Genomics (2016) 17:449 DOI 10.1186/s12864-016-2804-1 RESEARCH ARTICLE Open Access Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian1, Stephane Panserat1, Mathilde Dupont-Nivet2, Edwige Quillet2, Jerome Montfort3, Aurelie Le Cam3, Francoise Medale1, Sadasivam J. Kaushik1 and Inge Geurden1* Abstract Background: The achievement of sustainable feeding practices in aquaculture by reducing the reliance on wild-captured fish, via replacement of fish-based feed with plant-based feed, is impeded by the poor growth response seen in fish fed high levels of plant ingredients. -

Targeted Erythropoietin Selectively Stimulates Red Blood Cell Expansion in Vivo
Targeted erythropoietin selectively stimulates red blood cell expansion in vivo Devin R. Burrilla, Andyna Verneta, James J. Collinsa,b,c,d, Pamela A. Silvera,e,1, and Jeffrey C. Waya aWyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115; bSynthetic Biology Center, Massachusetts Institute of Technology, Cambridge, MA 02139; cInstitute for Medical Engineering & Science, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139; dBroad Institute of MIT and Harvard, Cambridge, MA 02139; and eDepartment of Systems Biology, Harvard Medical School, Boston, MA 02115 Edited by Ronald A. DePinho, University of Texas MD Anderson Cancer Center, Houston, TX, and approved March 30, 2016 (received for review December 23, 2015) The design of cell-targeted protein therapeutics can be informed peptide linker that permits simultaneous binding of both elements by natural protein–protein interactions that use cooperative phys- to the same cell surface. The targeting element anchors the mu- ical contacts to achieve cell type specificity. Here we applied this tated activity element to the desired cell surface (Fig. 1A, Middle), approach in vivo to the anemia drug erythropoietin (EPO), to direct thereby creating a high local concentration and driving receptor its activity to EPO receptors (EPO-Rs) on red blood cell (RBC) pre- binding despite the mutation (Fig. 1A, Bottom). Off-target sig- cursors and prevent interaction with EPO-Rs on nonerythroid cells, naling should be minimal (Fig. 1B) and should decrease in pro- such as platelets. Our engineered EPO molecule was mutated to portion to the mutation strength. weaken its affinity for EPO-R, but its avidity for RBC precursors Here we tested the chimeric activator strategy in vivo using was rescued via tethering to an antibody fragment that specifi- erythropoietin (EPO) as the drug to be targeted. -
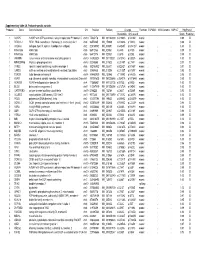
Supplementary Table 2A. Proband-Specific Variants Proband
Supplementary table 2A. Proband-specific variants Proband Gene Gene full name Chr Position RefSeqChange Function ESP6500 1000 Genome SNP137 PolyPhen2 Nucleotide Amino acid Score Prediction 1 AGAP5 ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 chr10 75435178 NM_001144000 c.G1240A p.V414M exonic . 0.99 D 1 REXO1L1 REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1 chr8 86574465 NM_172239 c.A1262G p.Y421C exonic . 0.98 D 1 COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) chr2 228169782 NM_000091 c.G4235T p.G1412V exonic . 1.00 D 1 KIAA1586 KIAA1586 chr6 56912133 NM_020931 c.C44G p.A15G exonic . 0.99 D 1 KIAA1586 KIAA1586 chr6 56912176 NM_020931 c.C87G p.D29E exonic . 0.99 D 1 JAKMIP3 Janus kinase and microtubule interacting protein 3 chr10 133955524 NM_001105521 c.G1574C p.G525A exonic . 1.00 D 1 MPHOSPH8 M-phase phosphoprotein 8 chr13 20240685 NM_017520 c.C2140T p.L714F exonic . 0.97 D 1 SYNE1 spectrin repeat containing, nuclear envelope 1 chr6 152783902 NM_033071 c.A2242T p.N748Y exonic . 0.97 D 1 CAND2 cullin-associated and neddylation-dissociated 2 (putative) chr3 12858835 NM_012298 c.C2125T p.H709Y exonic . 0.95 D 1 TDRD9 tudor domain containing 9 chr14 104460909 NM_153046 c.T1289G p.V430G exonic . 0.96 D 2 ASPM asp (abnormal spindle) homolog, microcephaly associated (Drosophila)chr1 197091625 NM_001206846 c.G3491A p.R1164H exonic . 0.99 D 2 ADAM29 ADAM metallopeptidase domain 29 chr4 175896951 NM_001130705 c.A275G p.D92G exonic . 1.00 D 2 BCO2 beta-carotene oxygenase 2 chr11 112087019 NM_001256398 c.G1373A p.G458E exonic . -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Expression Profiling of KLF4
Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in wild type MEFs. 98 Am J Cancer Res 2011;1(1):85-97 Table S1: Functional Annotation Clustering of Genes Up-Regulated in Klf4 -Null MEFs ILLUMINA_ID Gene Symbol Gene Name (Description) P -value Fold-Change Cell Cycle 8.00E-03 ILMN_1217331 Mcm6 MINICHROMOSOME MAINTENANCE DEFICIENT 6 40.36 ILMN_2723931 E2f6 E2F TRANSCRIPTION FACTOR 6 26.8 ILMN_2724570 Mapk12 MITOGEN-ACTIVATED PROTEIN KINASE 12 22.19 ILMN_1218470 Cdk2 CYCLIN-DEPENDENT KINASE 2 9.32 ILMN_1234909 Tipin TIMELESS INTERACTING PROTEIN 5.3 ILMN_1212692 Mapk13 SAPK/ERK/KINASE 4 4.96 ILMN_2666690 Cul7 CULLIN 7 2.23 ILMN_2681776 Mapk6 MITOGEN ACTIVATED PROTEIN KINASE 4 2.11 ILMN_2652909 Ddit3 DNA-DAMAGE INDUCIBLE TRANSCRIPT 3 2.07 ILMN_2742152 Gadd45a GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE 45 ALPHA 1.92 ILMN_1212787 Pttg1 PITUITARY TUMOR-TRANSFORMING 1 1.8 ILMN_1216721 Cdk5 CYCLIN-DEPENDENT KINASE 5 1.78 ILMN_1227009 Gas2l1 GROWTH ARREST-SPECIFIC 2 LIKE 1 1.74 ILMN_2663009 Rassf5 RAS ASSOCIATION (RALGDS/AF-6) DOMAIN FAMILY 5 1.64 ILMN_1220454 Anapc13 ANAPHASE PROMOTING COMPLEX SUBUNIT 13 1.61 ILMN_1216213 Incenp INNER CENTROMERE PROTEIN 1.56 ILMN_1256301 Rcc2 REGULATOR OF CHROMOSOME CONDENSATION 2 1.53 Extracellular Matrix 5.80E-06 ILMN_2735184 Col18a1 PROCOLLAGEN, TYPE XVIII, ALPHA 1 51.5 ILMN_1223997 Crtap CARTILAGE ASSOCIATED PROTEIN 32.74 ILMN_2753809 Mmp3 MATRIX METALLOPEPTIDASE -

Proquest Dissertations
INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. Each original is also photographed in one exposure and is included in reduced form at the back of the book. Photographs included in the original manuscript have been reproduced xerographically in this copy. Higher quality 6” x 9” black and white photographic prints are available for any photographs or illustrations appearing in this copy for an additional charge. Contact UMI directly to order. UMI Bell & Howell Information and Learning 300 North Zeeb Road, Ann Arbor, Ml 48106-1346 USA 800-521-0600 NOTE TO USERS Page(s) missing in number only; text follows. Microfilmed as received. 222-229 This reproduction is the best copy available. UMI MOLECULAR GENETIC AND BIOCHEMICAL STUDIES OF THE HUMAN AND MOUSE MHC COMPLEMENT GENE CLUSTERS DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Zhenyu Yang, M.S. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

When 7 Transmembrane Receptors Are Not G Protein–Coupled Receptors
When 7 transmembrane receptors are not G protein–coupled receptors Keshava Rajagopal, … , Robert J. Lefkowitz, Howard A. Rockman J Clin Invest. 2005;115(11):2971-2974. https://doi.org/10.1172/JCI26950. Commentary Classically, 7 transmembrane receptors transduce extracellular signals by coupling to heterotrimeric G proteins, although recent in vitro studies have clearly demonstrated that they can also signal via G protein–independent mechanisms. However, the physiologic consequences of this unconventional signaling, particularly in vivo, have not been explored. In this issue of the JCI, Zhai et al. demonstrate in vivo effects of G protein–independent signaling by the angiotensin II type 1 receptor (AT1R). In studies of the mouse heart, they compare the physiologic and biochemical consequences of transgenic cardiac-specific overexpression of a mutant AT1R incapable of G protein coupling with those of a wild-type receptor. Their results not only provide the first glimpse of the physiologic effects of this newly appreciated mode of signaling but also provide important and previously unappreciated clues as to the underlying molecular mechanisms. Find the latest version: https://jci.me/26950/pdf commentaries gene, spastin, regulates microtubule stability to 16. Wittmann, C.W., et al. 2001. Tauopathy in Dro- 21. Chan, Y.B., et al. 2003. Neuromuscular defects in modulate synaptic structure and function. Curr. sophila: neurodegeneration without neurofibril- a Drosophila survival motor neuron gene mutant. Biol. 14:1135–1147. lary tangles. Science. 293:711–714. Hum. Mol. Genet. 12:1367–1376. 12. Sherwood, N.T., Sun, Q., Xue, M., Zhang, B., and 17. Shapiro, W.R., and Young, D.F. -

R&D Assay for Alzheimer's Disease
R&DR&D assayassay forfor Alzheimer’sAlzheimer’s diseasedisease Target screening⳼ Ⲽ㬔 antibody array, ᢜ⭉㬔 ⸽ἐⴐ Amyloid β-peptide Alzheimer’s disease⯸ ኸᷠ᧔ ᆹ⸽ inhibitor, antibody, ELISA kit Surwhrph#Surilohu#Dqwlerg|#Duud| 6OUSFBUFE 1."5SFBUFE )41 $3&# &3, &3, )41 $3&# &3, &3, 壤伡庰䋸TBNQMF ɅH 侴䋸嵄䍴䋸BOBMZUFT䋸䬱娴哜塵 1$ 1$ 1$ 1$ 5IFNPTUSFGFSFODFEBSSBZT 1$ 1$ QQ α 34, .4, 503 Q α 34, .4, 503 %SVHTDSFFOJOH0òUBSHFUFòFDUT0ATHWAY涭廐 6OUSFBUFE 堄币䋸4BNQMF侴䋸8FTUFSOPS&-*4"䍘䧽 1."5SFBUFE P 8FTUFSOCMPU廽喜儤应侴䋸0, Z 4VCTUSBUF -JHIU )31DPOKVHBUFE1BO "OUJQIPTQIPUZSPTJOF .FBO1JYFM%FOTJUZ Y $BQUVSF"OUJCPEZ 5BSHFU"OBMZUF "SSBZ.FNCSBOF $3&# &3, &3, )41 .4, Q α 34, 503 Human XL Cytokine Array kit (ARY022, 102 analytes) Adiponectin,Aggrecan,Angiogenin,Angiopoietin-1,Angiopoietin-2,BAFF,BDNF,Complement,Component C5/C5a,CD14,CD30,CD40L, Chitinase 3-like 1,Complement Factor D,C-Reactive Protein,Cripto-1,Cystatin C,Dkk-1,DPPIV,EGF,EMMPRIN,ENA-78,Endoglin, Fas L,FGF basic,FGF- 7,FGF-19,Flt-3 L,G-CSF,GDF-15,GM-CSF,GRO-α,Grow th Hormone,HGF,ICAM-1,IFN-γ,IGFBP-2,IGFBP-3, IL-1α,IL-1β, IL-1ra,IL-2,IL-3,IL-4,IL- 5,IL-6,IL-8, IL-10,IL-11,IL-12, IL-13,IL-15,IL-16,IL-17A,IL-18 BPa,IL-19,IL-22, IL-23,IL-24,IL-27, IL-31,IL-32α/β/γ,IL-33,IL-34,IP-10,I-TAC,Kallikrein 3,Leptin,LIF,Lipocalin-2,MCP-1,MCP-3,M-CSF,MIF,MIG,MIP-1α/MIP-1β,MIP-3α,MIP-3β,MMP-9, Myeloperoxidase,Osteopontin, p70, PDGF-AA, PDGF-AB/BB,Pentraxin-3, PF4, RAGE, RANTES,RBP4,Relaxin-2, Resistin,SDF-1α,Serpin E1, SHBG, ST2, TARC,TFF3,TfR,TGF- ,Thrombospondin-1,TNF-α, uPAR, VEGF, Vitamin D BP Human Protease (34 analytes) /