Hereditary Angioedema: How to Approach It at the Emergency Department?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

R&D Assay for Alzheimer's Disease
R&DR&D assayassay forfor Alzheimer’sAlzheimer’s diseasedisease Target screening⳼ Ⲽ㬔 antibody array, ᢜ⭉㬔 ⸽ἐⴐ Amyloid β-peptide Alzheimer’s disease⯸ ኸᷠ᧔ ᆹ⸽ inhibitor, antibody, ELISA kit Surwhrph#Surilohu#Dqwlerg|#Duud| 6OUSFBUFE 1."5SFBUFE )41 $3&# &3, &3, )41 $3&# &3, &3, 壤伡庰䋸TBNQMF ɅH 侴䋸嵄䍴䋸BOBMZUFT䋸䬱娴哜塵 1$ 1$ 1$ 1$ 5IFNPTUSFGFSFODFEBSSBZT 1$ 1$ QQ α 34, .4, 503 Q α 34, .4, 503 %SVHTDSFFOJOH0òUBSHFUFòFDUT0ATHWAY涭廐 6OUSFBUFE 堄币䋸4BNQMF侴䋸8FTUFSOPS&-*4"䍘䧽 1."5SFBUFE P 8FTUFSOCMPU廽喜儤应侴䋸0, Z 4VCTUSBUF -JHIU )31DPOKVHBUFE1BO "OUJQIPTQIPUZSPTJOF .FBO1JYFM%FOTJUZ Y $BQUVSF"OUJCPEZ 5BSHFU"OBMZUF "SSBZ.FNCSBOF $3&# &3, &3, )41 .4, Q α 34, 503 Human XL Cytokine Array kit (ARY022, 102 analytes) Adiponectin,Aggrecan,Angiogenin,Angiopoietin-1,Angiopoietin-2,BAFF,BDNF,Complement,Component C5/C5a,CD14,CD30,CD40L, Chitinase 3-like 1,Complement Factor D,C-Reactive Protein,Cripto-1,Cystatin C,Dkk-1,DPPIV,EGF,EMMPRIN,ENA-78,Endoglin, Fas L,FGF basic,FGF- 7,FGF-19,Flt-3 L,G-CSF,GDF-15,GM-CSF,GRO-α,Grow th Hormone,HGF,ICAM-1,IFN-γ,IGFBP-2,IGFBP-3, IL-1α,IL-1β, IL-1ra,IL-2,IL-3,IL-4,IL- 5,IL-6,IL-8, IL-10,IL-11,IL-12, IL-13,IL-15,IL-16,IL-17A,IL-18 BPa,IL-19,IL-22, IL-23,IL-24,IL-27, IL-31,IL-32α/β/γ,IL-33,IL-34,IP-10,I-TAC,Kallikrein 3,Leptin,LIF,Lipocalin-2,MCP-1,MCP-3,M-CSF,MIF,MIG,MIP-1α/MIP-1β,MIP-3α,MIP-3β,MMP-9, Myeloperoxidase,Osteopontin, p70, PDGF-AA, PDGF-AB/BB,Pentraxin-3, PF4, RAGE, RANTES,RBP4,Relaxin-2, Resistin,SDF-1α,Serpin E1, SHBG, ST2, TARC,TFF3,TfR,TGF- ,Thrombospondin-1,TNF-α, uPAR, VEGF, Vitamin D BP Human Protease (34 analytes) / -

Kallikrein 13: a New Player in Coronaviral Infections
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.01.971499; this version posted March 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Kallikrein 13: a new player in coronaviral infections. 2 3 Aleksandra Milewska1,2, Katherine Falkowski2, Magdalena Kalinska3, Ewa Bielecka3, 4 Antonina Naskalska1, Pawel Mak4, Adam Lesner5, Marek Ochman6, Maciej Urlik6, Jan 5 Potempa2,7, Tomasz Kantyka3,8, Krzysztof Pyrc1,* 6 7 1 Virogenetics Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian 8 University, Gronostajowa 7a, 30-387 Krakow, Poland. 9 2 Microbiology Department, Faculty of Biochemistry, Biophysics and Biotechnology, 10 Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland. 11 3 Laboratory of Proteolysis and Post-translational Modification of Proteins, Malopolska 12 Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, 13 Poland. 14 4 Department of Analytical Biochemistry, Faculty of Biochemistry, Biophysics and 15 Biotechnology, Jagiellonian University, Gronostajowa 7 St., 30-387, Krakow, Poland. 16 5 University of Gdansk, Faculty of Chemistry, Wita Stwosza 63, 80-308 Gdansk, Poland. 17 6 Department of Cardiac, Vascular and Endovascular Surgery and Transplantology, Medical 18 University of Silesia in Katowice, Silesian Centre for Heart Diseases, Zabrze, Poland. 19 7 Centre for Oral Health and Systemic Diseases, University of Louisville School of Dentistry, 20 Louisville, KY 40202, USA. 21 8 Broegelmann Research Laboratory, Department of Clinical Science, University of Bergen, 22 5020 Bergen, Norway 23 24 25 26 27 28 29 30 31 * Correspondence should be addressed to Krzysztof Pyrc ([email protected]), Virogenetics 32 Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian University, 33 Gronostajowa 7, 30-387 Krakow, Poland; Phone number: +48 12 664 61 21; www: 34 http://virogenetics.info/. -

Proteome Profiler Human Protease Array Kit
Proteome ProfilerTM Array Human Protease Array Kit Catalog Number ARY021 For the parallel determination of the relative levels of selected human proteases. This package insert must be read in its entirety before using this product. For research use only. Not for use in diagnostic procedures. TABLE OF CONTENTS SECTION PAGE INTRODUCTION .....................................................................................................................................................................1 PRINCIPLE OF THE ASSAY ...................................................................................................................................................1 TECHNICAL HINTS .................................................................................................................................................................1 MATERIALS PROVIDED & STORAGE CONDITIONS ...................................................................................................2 OTHER SUPPLIES REQUIRED .............................................................................................................................................3 SUPPLIES REQUIRED FOR CELL LYSATE SAMPLES ...................................................................................................3 SUPPLIES REQUIRED FOR TISSUE LYSATE SAMPLES ...............................................................................................3 SAMPLE COLLECTION & STORAGE .................................................................................................................................4 -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Prostatic Trypsin-Like Kallikrein-Related Peptidases
Article in press - uncorrected proof Biol. Chem., Vol. 389, pp. 653–668, June 2008 • Copyright ᮊ by Walter de Gruyter • Berlin • New York. DOI 10.1515/BC.2008.078 Review Prostatic trypsin-like kallikrein-related peptidases (KLKs) and other prostate-expressed tryptic proteinases as regulators of signalling via proteinase-activated receptors (PARs) Andrew J. Ramsay1, Janet C. Reid1, Mark N. cesses by selective cleavage of specific substrates to Adams1, Hemamali Samaratunga2, Ying Dong1, influence cell behaviour (Turk, 2006). Well known physio- Judith A. Clements1 and John D. Hooper1,* logical examples include the proteinases of the blood coagulation (e.g., thrombin) (Davie et al., 1991), digestive 1 Institute of Health and Biomedical Innovation, (e.g., trypsin) (Yamashina, 1956) and wound healing (e.g., Queensland University of Technology, 6 Musk Avenue, plasmin) (Castellino and Ploplis, 2005) cascades. It is Kelvin Grove, Queensland 4059, Australia also clear that dysregulated serine proteinase activity 2 Department of Anatomical Pathology, Sullivan facilitates progression of various diseases including can- Nicolaides Pathology, Taringa 4068, Australia cer (Dano et al., 2005; Turk, 2006; Szabo and Bugge, * Corresponding author 2008). The cleavage site specificity of these enzymes is e-mail: [email protected] indicated by the residue six amino acids before the cat- alytic serine. In the active conformation of the proteinase, this residue is located at the bottom of a pocket in which Abstract the side-chain of the scissile bond of the substrate is inserted. An aspartate or, rarely, a glutamate at this site The prostate is a site of high expression of serine pro- indicates that the serine proteinase will preferentially teinases including members of the kallikrein-related cleave after substrate arginine or lysine residues (trypsin- peptidase (KLK) family, as well as other secreted and like or tryptic substrate specificity); a small hydrophobic membrane-anchored serine proteinases. -

Activation Profiles and Regulatory Cascades of the Human Kallikrein-Related Peptidases Hyesook Yoon
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2008 Activation Profiles and Regulatory Cascades of the Human Kallikrein-Related Peptidases Hyesook Yoon Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES ACTIVATION PROFILES AND REGULATORY CASCADES OF THE HUMAN KALLIKREIN-RELATED PEPTIDASES By HYESOOK YOON A Dissertation submitted to the Department of Chemistry and Biochemistry in partial fulfillment of the requirements for the degree of Doctor of Philosophy Degree Awarded: Fall Semester, 2008 The members of the Committee approve the dissertation of Hyesook Yoon defended on July 10th, 2008. ________________________ Michael Blaber Professor Directing Dissertation ________________________ Hengli Tang Outside Committee Member ________________________ Brian Miller Committee Member ________________________ Oliver Steinbock Committee Member Approved: ____________________________________________________________ Joseph B. Schlenoff, Chair, Department of Chemistry and Biochemistry The Office of Graduate Studies has verified and approved the above named committee members. ii ACKNOWLEDGMENTS I would like to dedicate this dissertation to my parents for all your support, and my sister and brother. I would also like to give great thank my advisor, Dr. Blaber for his patience, guidance. Without him, I could never make this achievement. I would like to thank to all the members in Blaber lab. They are just like family to me and I deeply appreciate their kindness, consideration and supports. I specially like to thank to Mrs. Sachiko Blaber for her endless guidance and encouragement. I would like to thank Dr Jihun Lee, Margaret Seavy, Rani and Doris Terry for helpful discussions and supports. -

Somamer Reagents Generated to Human Proteins Number Somamer Seqid Analyte Name Uniprot ID 1 5227-60
SOMAmer Reagents Generated to Human Proteins The exact content of any pre-specified menu offered by SomaLogic may be altered on an ongoing basis, including the addition of SOMAmer reagents as they are created, and the removal of others if deemed necessary, as we continue to improve the performance of the SOMAscan assay. However, the client will know the exact content at the time of study contracting. SomaLogic reserves the right to alter the menu at any time in its sole discretion. Number SOMAmer SeqID Analyte Name UniProt ID 1 5227-60 [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 1, mitochondrial Q15118 2 14156-33 14-3-3 protein beta/alpha P31946 3 14157-21 14-3-3 protein epsilon P62258 P31946, P62258, P61981, Q04917, 4 4179-57 14-3-3 protein family P27348, P63104, P31947 5 4829-43 14-3-3 protein sigma P31947 6 7625-27 14-3-3 protein theta P27348 7 5858-6 14-3-3 protein zeta/delta P63104 8 4995-16 15-hydroxyprostaglandin dehydrogenase [NAD(+)] P15428 9 4563-61 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 P19174 10 10361-25 2'-5'-oligoadenylate synthase 1 P00973 11 3898-5 26S proteasome non-ATPase regulatory subunit 7 P51665 12 5230-99 3-hydroxy-3-methylglutaryl-coenzyme A reductase P04035 13 4217-49 3-hydroxyacyl-CoA dehydrogenase type-2 Q99714 14 5861-78 3-hydroxyanthranilate 3,4-dioxygenase P46952 15 4693-72 3-hydroxyisobutyrate dehydrogenase, mitochondrial P31937 16 4460-8 3-phosphoinositide-dependent protein kinase 1 O15530 17 5026-66 40S ribosomal protein S3 P23396 18 5484-63 40S ribosomal protein -
![Kallikrein 13 Antibody Pair [HRP] NBP2-79509 Manual](https://docslib.b-cdn.net/cover/3877/kallikrein-13-antibody-pair-hrp-nbp2-79509-manual-2613877.webp)
Kallikrein 13 Antibody Pair [HRP] NBP2-79509 Manual
PRODUCT INFORMATION & ELISA MANUAL Kallikrein 13 Antibody Pair [HRP] NBP2-79509 Matched Antibody Pair utilized in an Enzyme- linked Immunosorbent Assay for quantitative detection of Human Kallikrein 13. For research use only. Not for diagnostic or therapeutic procedures. www.novusbio.com - P: 303.730.1950 - P: 888.506.6887 - F: 303.730.1966 - [email protected] Novus kits are guaranteed for 6 months from date of receipt BACKGROUND Human tissue kallikrein 13 (hK13), also known as KLK-L4 (kallikrein-like gene 4), is a member of the human tissue kallikrein family of serine proteases having diverse physiological functions in many tissues. The KLK13 gene resides on chromosome 19q13.3-4 along with other 14 members in a gene cluster and shares a high degree of homology. KLK13 is a trypsin-like, secreted serine protease expressed specifically in the testicular tissue including prostate, salivary gland, breast, and testis. Growing evidence suggests that many kallikreins are implicated in carcinogenesis and may play a role in metastasis. KLK13 may be involved in the pathogenesis and/or progression of breast and ovary cancers, and is regarded as a novel cancer biomarker. In addition, KLK13 interacts and forms complexes with several serum protease inhibitors, such as alpha2-macroglobulin, and its expression is regulated by steroid hormones. 2 PRINCIPLE OF THE TEST The Novus Biologicals Kallikrein 13 Antibody Pair [HRP] is asolid phase sandwich ELISA (Enzyme- Linked ImmunosorbentAssay). It utilizes a monoclonal antibody specific for Kallikrein 13 coated on a 96- well plate. Standards and samples areadded to the wells, and any Kallikrein 13 presentbinds to the immobilized antibody. -

Natural and Engineered Kallikrein Inhibitors: an Emerging Pharmacopoeia
Article in press - uncorrected proof Biol. Chem., Vol. 391, pp. 357–374, April 2010 • Copyright ᮊ by Walter de Gruyter • Berlin • New York. DOI 10.1515/BC.2010.037 Review Natural and engineered kallikrein inhibitors: an emerging pharmacopoeia Joakim E. Swedberg, Simon J. de Veer and ulated activation cascades, suggesting an involvement in a Jonathan M. Harris* diverse range of physiological processes (Pampalakis and Sotiropoulou, 2007). Both liver-derived KLKB1 and tissue- Institute of Health and Biomedical Innovation, Queensland derived KLK1, as well as KLK2 and KLK12 in vitro (Giusti University of Technology, Brisbane, Queensland 4059, et al., 2005), participate in the progressive activation of Australia bradykinin, a bioactive peptide involved in blood pressure * Corresponding author homeostasis and inflammation initiation (Bhoola et al., e-mail: [email protected] 1992). Although this is the only demonstration of classical kininogenic activity that was the original hallmark of kallik- Abstract rein proteases, the contribution of subsets of KLKs to vital physiological processes is well appreciated. Prostate- The kallikreins and kallikrein-related peptidases are serine expressed KLK2, 3, 4, 5 and 14 are involved in seminogelin proteases that control a plethora of developmental and home- hydrolysis (Lilja, 1985; Deperthes et al., 1996; Takayama et ostatic phenomena, ranging from semen liquefaction to skin al., 2001b; Michael et al., 2006; Emami and Diamandis, desquamation and blood pressure. The diversity of roles 2008), KLK6 and 8 have reported functions in defining neu- played by kallikreins has stimulated considerable interest in ral plasticity (Shimizu et al., 1998; Scarisbrick et al., 2002; these enzymes from the perspective of diagnostics and drug Tamura et al., 2006; Ishikawa et al., 2008) and KLK5, 7, 8 design. -

Natural and Synthetic Inhibitors of Kallikrein-Related Peptidases (Klks)
Biochimie 92 (2010) 1546e1567 Contents lists available at ScienceDirect Biochimie journal homepage: www.elsevier.com/locate/biochi Review Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs) Peter Goettig a,*, Viktor Magdolen b, Hans Brandstetter a a Division of Structural Biology, Department of Molecular Biology, University of Salzburg, Billrothstrasse 11, 5020 Salzburg, Austria b Klinische Forschergruppe der Frauenklinik, Klinikum rechts der Isar der TU München, Ismaninger Strasse 22, 81675 München, Germany article info abstract Article history: Including the true tissue kallikrein KLK1, kallikrein-related peptidases (KLKs) represent a family of fifteen Received 24 February 2010 mammalian serine proteases. While the physiological roles of several KLKs have been at least partially Accepted 29 June 2010 elucidated, their activation and regulation remain largely unclear. This obscurity may be related to the fact Available online 6 July 2010 that a given KLK fulfills many different tasks in diverse fetal and adult tissues, and consequently, the timescale of some of their physiological actions varies significantly. To date, a variety of endogenous þ Keywords: inhibitors that target distinct KLKs have been identified. Among them are the attenuating Zn2 ions, Tissue kallikrein fi active site-directed proteinaceous inhibitors, such as serpins and the Kazal-type inhibitors, or the huge, Speci city pockets fi Inhibitory compound unspeci c compartment forming a2-macroglobulin. Failure of these inhibitory systems can lead to certain Zinc pathophysiological conditions. One of the most prominent examples is the Netherton syndrome, which is Rule of five caused by dysfunctional domains of the Kazal-type inhibitor LEKTI-1 which fail to appropriately regulate KLKs in the skin. -
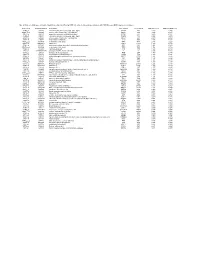
Table 5. Probesets (54) Representing the Top 50 Transcripts Identified by PAM That Delineate Lung Adenocarcinomas with NED from Non-NED Lung Adenocarcinomas
Table 5. Probesets (54) representing the top 50 transcripts identified by PAM that delineate lung adenocarcinomas with NED from non-NED lung adenocarcinomas Probeset ID GenBank/TIGR ID Gene Name Gene Symbol LocusLink ID PAM NED score PAM non-NED score 36299_at X02330 calcitonin/calcitonin-related polypeptide, alpha CALCA 796 3.810 -0.405 40544_g_at L08424 achaete-scute complex-like 1 (Drosophila) ASCL1 429 3.543 -0.376 40649_at X64810 proprotein convertase subtilisin/kexin type 1 PCSK1 5122 3.076 -0.327 36300_at X15943 calcitonin/calcitonin-related polypeptide, alpha CALCA 796 2.628 -0.279 40543_at L08424 achaete-scute complex-like 1 (Drosophila) ASCL1 429 2.470 -0.262 31477_at L08044 trefoil factor 3 (intestinal) TFF3 7033 2.175 -0.231 37898_r_at AI985964 trefoil factor 3 (intestinal) TFF3 7033 2.146 -0.228 40035_at AB012917 kallikrein 11 KLK11 11012 1.878 -0.199 40201_at M76180 dopa decarboxylase (aromatic L-amino acid decarboxylase) DDC 1644 1.864 -0.198 37897_s_at AI985964 trefoil factor 3 (intestinal) TFF3 7033 1.748 -0.186 36606_at X51405 carboxypeptidase E CPE 1363 1.543 -0.164 1745_at HG4679-HT5104 Oncogene Ret/Ptc, Fusion Activated --- --- 1.525 -0.162 37019_at J00129 fibrinogen, B beta polypeptide FGB 2244 1.459 -0.155 33426_at Y00064 chromogranin B (secretogranin 1) CHGB 1114 1.383 -0.147 35966_at X71125 glutaminyl-peptide cyclotransferase (glutaminyl cyclase) QPCT 25797 1.341 -0.142 35415_at X12901 villin 1 VIL1 7429 1.335 -0.142 35343_at M37400 glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) GOT1