Signal Transduction Discover
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Overexpresssion of Tyrosine Kinase Discoidin Domain Receptor I (DDR1 ) in Transitional Cell Carcinoma
Overexpresssion of Tyrosine Kinase Discoidin domain receptor I (DDR1 ) in Transitional cell Carcinoma Szu-Ting Chen,* and Shie-Liang Hsieh* Institute and Department of Microbiology and Immunology, National Yang-Ming University, Taipei, Taiwan; To whom proofs are to be sent: Shie-Liang Hsieh, Department and Institute of Microbiology and Immunology, National Yang-Ming University, Shih-Pai, Taipei 112, Taiwan. E-mail address: [email protected] Telephone number: 886-2-28267161 Fax number: 886-2-28277933 INTRODUCTION DDR1, discoidin domain receptor 1, belongs to the novel subfamily of tyrosine kinase receptor, which forms homodimer upon ligand engagement. DDR1 is distinguished from other receptor tyrosine kinase by the discoidin domain in their extracellular domain, which is a homology region originally identified in Dictyostelium discoideum (slime mold) protein, discoidin I, and involves in cells aggregation. Discoidin-1 has binding specificity toward galactose and N-acetyl galactosamine and is essential for slime mold cells adhesion, migration and aggregation during its development, suggesting that DDR1 shared similar biologically function in mammalian [1-3]. DDR1 has been found mainly distributed and in human tissue epithelia, such as kidney, breast, lung [3], bronchial [4] and keratinocytes [5]. Furthermore, DDR1 has been also reported its expression in immune system like the monocyte-derived dendritic cells, annotated as CD167 [6] and macrophage[7]. However, recently, the overexpression of DDR1 has been detected in several human cancers, such as primary breast cancer [1, 8, 9] ovarian[10, 11], brain[12], esophageal cancer [13], and TCC (our data unpublished) in which raising the possibility that DDR1 may play a important role in tumorigenesis [14]. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Neprilysin Is Required for Angiotensin-(1-7)
Page 1 of 39 Diabetes NEPRILYSIN IS REQUIRED FOR ANGIOTENSIN-(1-7)’S ABILITY TO ENHANCE INSULIN SECRETION VIA ITS PROTEOLYTIC ACTIVITY TO GENERATE ANGIOTENSIN-(1-2) Gurkirat S. Brara, Breanne M. Barrowa, Matthew Watsonb, Ryan Griesbachc, Edwina Chounga, Andrew Welchc, Bela Ruzsicskad, Daniel P. Raleighb, Sakeneh Zraikaa,c aVeterans Affairs Puget Sound Health Care System, Seattle, WA 98108, United States bDepartment of Chemistry, Stony Brook University, Stony Brook, NY 11794, United States cDivision of Metabolism, Endocrinology and Nutrition, Department of Medicine, University of Washington, Seattle, WA 98195, United States dInstitute for Chemical Biology and Drug Discovery, Stony Brook University, Stony Brook, NY 11794, United States Short Title: Angiotensin-(1-7) and insulin secretion Word count: 3997; Figure count: 8 main (plus 3 Online Suppl.); Table count: 1 Online Suppl. Correspondence to: Sakeneh Zraika, PhD 1660 South Columbian Way (151) Seattle, WA, United States Tel: 206-768-5391 / Fax: 206-764-2164 Email: [email protected] 1 Diabetes Publish Ahead of Print, published online May 30, 2017 Diabetes Page 2 of 39 ABSTRACT Recent work has renewed interest in therapies targeting the renin-angiotensin system (RAS) to improve β-cell function in type 2 diabetes. Studies show that generation of angiotensin-(1-7) by angiotensin converting enzyme 2 (ACE2) and its binding to the Mas receptor (MasR) improves glucose homeostasis, partly by enhancing glucose-stimulated insulin secretion (GSIS). Thus, islet ACE2 upregulation is viewed as a desirable therapeutic goal. Here, we show that although endogenous islet ACE2 expression is sparse, its inhibition abrogates angiotensin-(1-7)-mediated GSIS. However, a more widely expressed islet peptidase, neprilysin, degrades angiotensin-(1-7) into several peptides. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

View a Copy of This Licence, Visit
Choi et al. BMC Cardiovascular Disorders (2020) 20:360 https://doi.org/10.1186/s12872-020-01636-5 RESEARCH ARTICLE Open Access Soluble neprilysin and long-term clinical outcomes in patients with coronary artery disease undergoing percutaneous coronary intervention: a retrospective cohort study Ik Jun Choi1, Sungmin Lim2* , Youngdeok Hwang3, Dongjae Lee1, Won Jik Lee1, Kwan Yong Lee1, Mi-Jeong Kim1 and Doo Soo Jeon1 Abstract Background: Neprilysin has an essential role in regulating fluid balance and vascular resistance, and neprilysin inhibitors have shown beneficial effects in patients with heart failure. However, the potential predictive value of neprilysin levels as a biomarker for cardiovascular risk remains unclear. The aim of this study was to assess the prognostic value of soluble neprilysin (sNEP) levels in patients with ischemic heart disease. Methods: Neprilysin levels were measured in 694 consecutive patients with coronary artery disease (CAD) undergoing percutaneous coronary intervention (PCI). These patients were classified into two groups according to their serum levels of neprilysin and categorized into the lower neprilysin group (n = 348) and the higher neprilysin group (n = 346). The primary clinical endpoint was all-cause mortality, and the secondary endpoint was a composite of major adverse cardiac events (MACE). Results: The median sNEP level was 76.0 pg/ml. The median sNEP levels were higher in patients with left ventricular ejection fraction (LVEF) ≥40% (77.6 pg/ml, interquartile range 46.6–141.3) than in those with LVEF < 40% (70.0 pg/ml, interquartile range 47.1–100.6; P = 0.032). Among all patients, each clinical outcome and MACE did not differ significantly according to the groups divided into median, tertile, or quartile of sNEP levels during a median follow-up of 28.4 months. -

Stromal CD10 Expression in Breast Cancer Correlates with Tumor Invasion and Cancer Stem Cell Phenotype
Louhichi et al. BMC Cancer (2018) 18:49 DOI 10.1186/s12885-017-3951-8 RESEARCH ARTICLE Open Access Stromal CD10 expression in breast cancer correlates with tumor invasion and cancer stem cell phenotype Tahani Louhichi, Hanene Saad, Myriam Ben Dhiab, Sonia Ziadi and Mounir Trimeche* Abstract Background: Previous investigations have indicated that CD10 is associated with biological aggressivity in human cancers, but the use of this marker for diagnosis and prognosis is more complex. The aim of this study was to evaluate the expression of CD10 in breast cancer and its association with the clinicopathological features. In addition, we investigated whether a relationship exists between CD10 expression and cancer stem cells. Methods: CD10 expression was examined by the immunohistochemistry in a series of 133 invasive breast carcinoma cases. Results were correlated to several clinicopathological parameters. Cancer stem cell phenotype was assessed by the immunohistochemical analysis of CD44 and ALDH1. Results: Significant CD10 expression was found in the fusiform stromal cells in 19.5% of the cases and in the neoplastic cells in 7% of the cases. The stromal CD10 positivity was more frequently found in tumors with lymph node metastasis (p = 0.01) and a high histological grade (p = 0.01). However, CD10 expression by the neoplastic cells correlates with a high histological grade (p = 0.03) and the absence of estrogen (p = 0.002) as well as progesterone (p = 0.001) receptor expression. We also found that CD10 expression by the stromal cells, but not by the neoplastic cells, correlates significantly with the expression of cancer stem cell markers (CD44+/ALDH1+) (p = 0.002). -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

Angiotensin Receptor Neprilysin Inhibition (ARNI) Following Acute Myocardial Infarction: Primary Results of the PARADISE-MI Trial Marc A
Angiotensin Receptor Neprilysin Inhibition (ARNI) Following Acute Myocardial Infarction: Primary Results of the PARADISE-MI Trial Marc A. Pfeffer, MD, PhD Distinguished Dzau Professor of Medicine Harvard Medical School Cardiovascular Division, Brigham and Women’s Hospital for the PARADISE-MI Committees, National Leaders and Investigators SAVE AIRE TRACE Radionuclide Clinical and/or Echocardiographic EF ≤ 40% radiographic signs EF ≤ 35% (1992) of HF (1993) (1995) 0.4 All-Cause Mortality 0.35 0.3 0.25 Placebo ACE-I 0.2 0.15 Placebo: 866/2971 (29.1%) Probability of of Probability Event 0.1 ACE-I: 702/2995 (23.4%) 0.05 OR: 0.74 (0.66–0.83) 0 Years 0 1 2 3 4 ACE-I 2995 2250 1617 892 223 Placebo 2971 2184 1521 853 138 Flather MD, et al. Lancet. 2000;355:1575–1581 Mortality in SAVE, TRACE, AIRE, and VALIANT Favors Active Drug Pfeffer,Pfeffer, McMurray, McMurray, Velazquez, Velazquez, et etal. al. N NEngl Engl J MedJ Med2003;3492003;349 2014 40 Enalapril 1117 32 (n=4212) 914 24 LCZ696 (n=4187) Meier Estimate of Meier Estimate 16 - HR = 0.80 (0.73-0.87) Cumulative (%) Rates Cumulative 8 Kaplan P = 0.0000002 Number needed to treat = 21 0 0 180 360 540 720 900 1080 1260 Patients at Risk Days After Randomization LCZ696 4187 3922 3663 3018 2257 1544 896 249 Enalapril 4212 3883 3579 2922 2123 1488 853 236 McMurray, N Engl J Med. 2014 AMI (0.5-7 days with LVEF ≤40% and/or pulmonary congestion) PLUS any risk enhancer Age ≥70 years Atrial fibrillation eGFR <60 LVEF < 30% Diabetes Killip class ≥III Prior MI STEMI without reperfusion Major Exclusions: Prior HF Clinical instability eGFR <30 Sacubitril/Valsartan Ramipril No run-in Target 97/103 mg BID Target 5 mg BID double-blind -controlled N=2830 active N=2831 Event driven: 711 primary endpoints Median follow-up: 23 months Primary Endpoint: CV death, HF hospitalization, outpatient development of HF Jering, Eur J ACC.21 Secondary Endpoint: CV death or first HF hospitalization Heart Fail. -

Mesenchymal–Epithelial Interactions Involving Epiregulin in Tuberous Sclerosis Complex Hamartomas
Mesenchymal–epithelial interactions involving epiregulin in tuberous sclerosis complex hamartomas Shaowei Li*, Fumiko Takeuchi*, Ji-an Wang*, Qingyuan Fan*, Toshi Komurasaki†, Eric M. Billings‡, Gustavo Pacheco-Rodriguez‡, Joel Moss‡, and Thomas N. Darling*§ *Department of Dermatology, Uniformed Services University of the Health Sciences, 4301 Jones Bridge Road, Bethesda, MD 20814-4712; †Molecular Biology Laboratory, Molecular and Pharmacology Laboratories, Taisho Pharmaceutical Co., Ltd., 430-1 Yoshino-cho, Saitma-shi, Saitama 331-9530, Japan; and ‡Translational Medicine Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Building 10, Room 6D05, MSC 1590, Bethesda, MD 20892-1590 Communicated by Martha Vaughan, National Institutes of Health, Bethesda, MD, December 31, 2007 (received for review November 30, 2007) Patients with tuberous sclerosis complex (TSC) develop hamarto- Like other hamartomas, those in TSC skin contain abnormal mas containing biallelic inactivating mutations in either TSC1 or numbers of several types of cells. In the dermis, there are TSC2, resulting in mammalian target of rapamycin (mTOR) activa- increased numbers of large stellate fibroblasts, capillaries, and tion. Hamartomas overgrow epithelial and mesenchymal cells in dermal dendritic cells (6–9). The epidermis is acanthotic (i.e., TSC skin. The pathogenetic mechanisms for these changes had not thickened from increased numbers of keratinocytes in the spi- been investigated, and the existence or location of cells with nous layer). Acanthosis is pronounced in PFs and variable in AFs biallelic mutations (‘‘two-hit’’ cells) was unclear. We compared TSC (7, 8). The epidermis of treated AFs, several months after argon skin hamartomas (angiofibromas and periungual fibromas) with or CO2 laser surgery, no longer appears acanthotic (10, 11). -
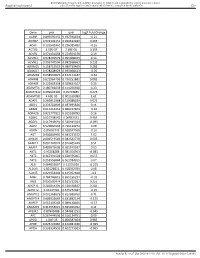
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Inhibition of DDR1-BCR Signalling by Nilotinib As a New Therapeutic
Inhibition of DDR1-BCR signalling by nilotinib as a new therapeutic strategy for metastatic colorectal cancer Maya Jeitany, Cédric Leroy, Priscillia Tosti, Marie Lafitte, Jordy Le Guet, Valérie Simon, Debora Bonenfant, Bruno Robert, Fanny Grillet, Caroline Mollevi, et al. To cite this version: Maya Jeitany, Cédric Leroy, Priscillia Tosti, Marie Lafitte, Jordy Le Guet, et al.. Inhibition of DDR1- BCR signalling by nilotinib as a new therapeutic strategy for metastatic colorectal cancer. EMBO Molecular Medicine, Wiley Open Access, 2018, 10 (4), pp.e7918. 10.15252/emmm.201707918. hal- 01872978 HAL Id: hal-01872978 https://hal.archives-ouvertes.fr/hal-01872978 Submitted on 12 Jan 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Research Article Inhibition of DDR1-BCR signalling by nilotinib as a new therapeutic strategy for metastatic colorectal cancer Maya Jeitany1,†, Cédric Leroy1,2,3,†, Priscillia Tosti1,†, Marie Lafitte1, Jordy Le Guet1, Valérie Simon1, Debora Bonenfant2, Bruno Robert4, Fanny Grillet5, Caroline Mollevi4, Safia El Messaoudi4, Amaëlle Otandault4, Lucile Canterel-Thouennon4, Muriel Busson4, Alain R Thierry4, Pierre Martineau4, Julie Pannequin5, Serge Roche1,*,† & Audrey Sirvent1,†,** Abstract The current clinical management involves surgical removal of the primary tumour, often associated with chemotherapy.