Clinicogenomic Analysis of FGFR2-Rearranged Cholangiocarcinoma Identifies Correlates of Response and Mechanisms of Resistance to Pemigatinib
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Leukemia Insights June 2021
JUNE 2021 In this month’s Leukemia Insights newsletter, written by Prithviraj Bose, M.D. and Srdan Verstovsek, M.D., Ph.D., and sponsored in part by the Charif Souki Cancer Research ABOUT MyMDAnderson Fund, we discuss our novel therapeutic approaches for the rare hematologic malignancies, systemic mastocytosis and myeloid/lymphoid neoplasms with myMDAnderson is a secure, eosinophilia. Learn more about our Leukemia program. personalized web site helping community physicians expedite patient referrals, as well as improve continuity Spotlight on rare, atypical, myeloid of care through information access and streamlined communications. neoplasms: systemic mastocytosis and Physicians who have referred patients myeloid/lymphoid neoplasms with to MD Anderson or plan to do so, can utilize the HIPAA compliant features eosinophilia and FGFR1 rearrangements of myMDAnderson to: • Refer a patient • View your patient's appointments Systemic Mastocytosis Access patient reports • Send and receive secure Systemic mastocytosis (SM) is a rare myeloid neoplasm messages driven in approximately 95% of cases by an activating mutation in c-KIT, usually D816V. SM is characterized as indolent, smoldering or advanced, based on the presence JOIN THE COVERSATION and number of so-called B- and C-findings. The latter Connect with us. signify organ damage and are a hallmark of advanced SM (AdvSM). AdvSM, in turn, is typically sub-classified as aggressive SM (ASM), SM with an associated hematologic neoplasm (SM-AHN, the most common subtype) and mast cell leukemia (MCL). Patients with indolent SM (ISM) and smoldering SM (SSM) enjoy much better survival than JOIN OUR MAILING LIST those with AdvSM, although symptoms in all three subtypes can be severe and debilitating. -

Incyte Announces the European Commission Approval of Pemazyre
3/29/2021 Incyte Announces the European Commission Approval of Pemazyre® (pemigatinib) as a Treatment for Adults with Locally Advanced or Metastatic Cholangiocarcinoma with a Fibroblast Growth Factor Receptor 2 (FGFR2) Fusion or Rearrangement - Pemazyre is the rst targeted therapy approved in the EU for this indication WILMINGTON, Del.--(BUSINESS WIRE)-- Incyte (Nasdaq:INCY) today announced that the European Commission (EC) has approved Pemazyre® (pemigatinib) for the treatment of adults with locally advanced or metastatic cholangiocarcinoma with a broblast growth factor receptor 2 (FGFR2) fusion or rearrangement that have progressed after at least one prior line of systemic therapy. The decision follows the positive opinion received from the European Medicines Agency’s Committee for Medicinal Products for Human Use in January 2021 recommending the conditional marketing authorization of Pemazyre. “Pemazyre’s approval is a crucial milestone for patients with FGFR2 positive cholangiocarcinoma. It is the rst new treatment option to be made available to these patients in the EU in over a decade and has demonstrated a high rate of durable responses in a setting where historically there has been no eective standard of care,” said Hervé Hoppenot, Chief Executive Ocer, Incyte. “We now look forward to working with individual countries in Europe to ensure eligible patients can access this new treatment as soon as possible.” The EC decision is based on data from the FIGHT-202 study evaluating the safety and ecacy of Pemazyre in adult patients with previously treated, locally advanced or metastatic cholangiocarcinoma with documented FGF/FGFR status. Interim results from FIGHT-202 demonstrated that in patients harboring FGFR2 fusions or rearrangements (Cohort A [108 patients]), Pemazyre monotherapy resulted in an overall response rate (ORR) of 37 percent (primary endpoint) and a median duration of response (DOR) of 8 months (secondary endpoint) based on an independent central radiographic review. -
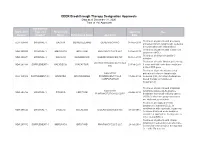
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities -

Rxoutlook® 1St Quarter 2019
® RxOutlook 1st Quarter 2020 optum.com/optumrx a RxOutlook 1st Quarter 2020 Orphan drugs continue to feature prominently in the drug development pipeline In 1983 the Orphan Drug Act was signed into law. Thirty seven years later, what was initially envisioned as a minor category of drugs has become a major part of the drug development pipeline. The Orphan Drug Act was passed by the United States Congress in 1983 in order to spur drug development for rare conditions with high unmet need. The legislation provided financial incentives to manufacturers if they could demonstrate that the target population for their drug consisted of fewer than 200,000 persons in the United States, or that there was no reasonable expectation that commercial sales would be sufficient to recoup the developmental costs associated with the drug. These “Orphan Drug” approvals have become increasingly common over the last two decades. In 2000, two of the 27 (7%) new drugs approved by the FDA had Orphan Designation, whereas in 2019, 20 of the 48 new drugs (42%) approved by the FDA had Orphan Designation. Since the passage of the Orphan Drug Act, 37 years ago, additional regulations and FDA designations have been implemented in an attempt to further expedite drug development for certain serious and life threatening conditions. Drugs with a Fast Track designation can use Phase 2 clinical trials to support FDA approval. Drugs with Breakthrough Therapy designation can use alternative clinical trial designs instead of the traditional randomized, double-blind, placebo-controlled trial. Additionally, drugs may be approved via the Accelerated Approval pathway using surrogate endpoints in clinical trials rather than clinical outcomes. -

Facts and New Hopes on Selective FGFR Inhibitors in Solid Tumors Francesco Facchinetti1, Antoine Hollebecque2, Rastislav Bahleda2, Yohann Loriot3, Ken A
Published OnlineFirst October 4, 2019; DOI: 10.1158/1078-0432.CCR-19-2035 CLINICAL CANCER RESEARCH | REVIEW Facts and New Hopes on Selective FGFR Inhibitors in Solid Tumors Francesco Facchinetti1, Antoine Hollebecque2, Rastislav Bahleda2, Yohann Loriot3, Ken A. Olaussen1, Christophe Massard2, and Luc Friboulet1 ABSTRACT ◥ Precision oncology relies on the identification of molecular altera- molecularly guided treatments. Matching molecularly selected tions, responsible for tumor initiation and growth, which are suitable tumors with selective FGFR inhibitors has indeed led to promising targets of specific inhibitors. The development of FGFR inhibitors results in phase I and II trials, justifying their registration to be represents an edifying example of the rapid evolution in the field of expected in a near future, such as the recent accelerated approval of targeted oncology, with 10 different FGFR tyrosine kinase inhibitors erdafitinib granted by the FDA for urothelial cancer. Widening our actually under clinical investigation. In parallel, the discovery of knowledge of the activity, efficacy, and toxicities relative to the FGFR activating molecular alterations (mainly FGFR3 mutations selective FGFR tyrosine kinase inhibitors under clinical investi- and FGFR2 fusions) across many tumor types, especially urothelial gation, according to the exact FGFR molecular alteration, will be carcinomas and intrahepatic cholangiocarcinomas, widens the selec- crucial to determine the optimal therapeutic strategy for patients tion of patients that might benefit from selective FGFR inhibitors. suffering from FGFR-driven tumors. Similarly, identifying with The ongoing concomitant clinical evaluation of selective FGFR appropriate molecular diagnostic, every single tumor harboring inhibitors in molecularly selected solid tumors brings new hopes targetable FGFR alterations will be of utmost importance to attain for patients with metastatic cancer, for tumors so far excluded from the best outcomes for patients with FGFR-driven cancer. -

Pemazyre – Criteria (PDF)
Criteria-Based Consultation Prescribing Program CRITERIA FOR DRUG COVERAGE Pemigatinib (Pemazyre) Notes: • Quantity Limits: Yes (#14 tablets per 21 days) Initiation (new start) criteria: Non-formulary pemigatinib (Pemazyre) will be covered on the prescription drug benefit when the following criteria are met: • Prescribed by a hematologist or oncologist AND • ECOG 0-1 AND • Age > 18 AND • Diagnosis of unresectable, locally advanced or metastatic cholangiosarcoma AND • Prior treatment with a first-line chemotherapy regimen per National Comprehensive Cancer Network (NCCN) Hepatobiliary Cancers Guideline (e.g. gemcitabine+cisplatin/oxaliplatin, 5- FU/capecitabine+cisplatin/oxaliplatin, gemcitabine+nab-paclitaxel, gemcitabine+capecitabine, gemcitabine+nab-paclitaxel+cisplatin, 5-FU, capecitabine, gemcitabine, pembrolizumab, entrectinib, larotrectinib) per NCCN Biliary Tract Cancers Guidelines AND • Fibroblast growth factor receptor 2 (FGFR2) fusion or rearrangement Criteria for current Kaiser Permanente members already taking the medication who have not been reviewed previously: Non-formulary pemigatinib (Pemazyre) will be covered on the prescription drug benefit when the following criteria are met: kp.org cps/awc Revised 05/13/21 Effective 07/15/21 Criteria-Based Consultation Prescribing Program CRITERIA FOR DRUG COVERAGE Pemigatinib (Pemazyre) • Prescribed by a hematologist or oncologist AND • Age > 18 AND • Diagnosis of unresectable, locally advanced or metastatic cholangiosarcoma AND • Fibroblast growth factor receptor 2 (FGFR2) genetic -

Tyrosine Kinase Inhibitors for Solid Tumors in the Past 20 Years (2001–2020) Liling Huang†, Shiyu Jiang† and Yuankai Shi*
Huang et al. J Hematol Oncol (2020) 13:143 https://doi.org/10.1186/s13045-020-00977-0 REVIEW Open Access Tyrosine kinase inhibitors for solid tumors in the past 20 years (2001–2020) Liling Huang†, Shiyu Jiang† and Yuankai Shi* Abstract Tyrosine kinases are implicated in tumorigenesis and progression, and have emerged as major targets for drug discovery. Tyrosine kinase inhibitors (TKIs) inhibit corresponding kinases from phosphorylating tyrosine residues of their substrates and then block the activation of downstream signaling pathways. Over the past 20 years, multiple robust and well-tolerated TKIs with single or multiple targets including EGFR, ALK, ROS1, HER2, NTRK, VEGFR, RET, MET, MEK, FGFR, PDGFR, and KIT have been developed, contributing to the realization of precision cancer medicine based on individual patient’s genetic alteration features. TKIs have dramatically improved patients’ survival and quality of life, and shifted treatment paradigm of various solid tumors. In this article, we summarized the developing history of TKIs for treatment of solid tumors, aiming to provide up-to-date evidence for clinical decision-making and insight for future studies. Keywords: Tyrosine kinase inhibitors, Solid tumors, Targeted therapy Introduction activation of tyrosine kinases due to mutations, translo- According to GLOBOCAN 2018, an estimated 18.1 cations, or amplifcations is implicated in tumorigenesis, million new cancer cases and 9.6 million cancer deaths progression, invasion, and metastasis of malignancies. occurred in 2018 worldwide [1]. Targeted agents are In addition, wild-type tyrosine kinases can also func- superior to traditional chemotherapeutic ones in selec- tion as critical nodes for pathway activation in cancer. -

Unitedhealthcare Cancer Therapy Pathways Program Regimens
UnitedHealthcare Cancer Therapy Pathways Program Overview 2 Breast Cancer 3 Pancreatic Cancer 15 Melanoma 19 Colon/Rectal Cancer 23 Hepatobiliary Cancers 30 Lung Cancer, Small Cell 35 Lung Cancer, Non-Small Cell 40 Ovarian, Fallopian and Primary Peritoneal Cancer 56 Head and Neck Cancer 67 Multiple Myeloma 74 Lymphoma, Diffuse Large B-Cell 86 Lymphoma, Follicular 92 Lymphoma, Marginal Zone 97 Lymphoma, Mantle Cell 101 Change control 1 OVERVIEW Cancer Therapy Pathways Program With the rapid approval of new therapies, along with the rising cost of cancer care, pathways serve a critical role in the delivery of high-quality and high-value cancer treatments while reducing an unwarranted variation in care. The UnitedHealthcare Cancer Therapy Pathways Program aims to improve quality and value in cancer care by identifying anti-cancer regimens supported by evidence-based guidelines to help reduce total cost of care and improve outcomes. The program’s regimens are selected on the basis of clinical benefit (efficacy) and side-effect profile (toxicity). Among regimens with comparable efficacy and toxicity, additional consideration is given to the frequency of hospitalizations during therapy, duration of therapy, drug costs and total cost of care. Care decisions are between the physician and the patient The Cancer Therapy Pathways Program is not a substitute for the experience and judgment of a physician or other health care professional. Any clinician participating in the program must use independent medical judgment in the context of individual -

Strengthening the Accelerated Approval Pathway: an Analysis of Potential Policy Reforms and Their Impact on Uncertainty, Access, Innovation, and Costs
STRENGTHENING THE ACCELERATED APPROVAL PATHWAY: AN ANALYSIS OF POTENTIAL POLICY REFORMS AND THEIR IMPACT ON UNCERTAINTY, ACCESS, INNOVATION, AND COSTS April 26, 2021 Anna Kaltenboeck, MA, MBA Program Director and Senior Health Economist, Center for Health Policy and Outcomes Memorial Sloan Kettering Cancer Center Amanda Mehlman Director of Strategic Partnerships Institute for Clinical and Economic Review Steven D. Pearson, MD, MSc President Institute for Clinical and Economic Review White Paper: Strengthening the Accelerated Approval Pathway 1 Table of Contents Introduction ........................................................................................................................................... 4 The Challenge ..................................................................................................................................... 4 Structure of This Paper ...................................................................................................................... 6 Methods ............................................................................................................................................. 7 Background ............................................................................................................................................ 8 The Accelerated Approval Pathway ................................................................................................... 8 What is Accelerated Approval? ..................................................................................................... -

Horizon Scanning Status Report December 2020
PCORI Health Care Horizon Scanning System Volume 2 Issue 4 Horizon Scanning Status Report December 2020 Prepared for: Patient-Centered Outcomes Research Institute 1828 L St., NW, Suite 900 Washington, DC 20036 Contract No. MSA-HORIZSCAN-ECRI-ENG-2018.7.12 Prepared by: ECRI Institute 5200 Butler Pike Plymouth Meeting, PA 19462 Investigators: Randy Hulshizer, MA, MS Damian Carlson, MS Christian Cuevas, PhD Andrea Druga, MSPAS, PA-C Marcus Lynch, PhD, MBA Misha Mehta, MS Prital Patel, MPH Brian Wilkinson, MA Donna Beales, MLIS Jennifer De Lurio, MS Eloise DeHaan, BS Eileen Erinoff, MSLIS Cassia Hulshizer, BBA Madison Kimball, MS Maria Middleton, MPH Melinda Rossi, BS Diane Robertson, BA Kelley Tipton, MPH Rosemary Walker, MLIS Andrew Furman, MD, MMM, FACEP Statement of Funding and Purpose This report incorporates data collected during implementation of the Patient-Centered Outcomes Research Institute (PCORI) Health Care Horizon Scanning System, operated by ECRI under contract to PCORI, Washington, DC (Contract No. MSA-HORIZSCAN-ECRI-ENG-2018.7.12). The findings and conclusions in this document are those of the authors, who are responsible for its content. No statement in this report should be construed as an official position of PCORI. An intervention that potentially meets inclusion criteria might not appear in this report simply because the Horizon Scanning System has not yet detected it or it does not yet meet the inclusion criteria outlined in the PCORI Health Care Horizon Scanning System: Horizon Scanning Protocol and Operations Manual. Inclusion or absence of interventions in the horizon scanning reports will change over time as new information is collected; therefore, inclusion or absence should not be construed as either an endorsement or a rejection of specific interventions. -

Pemigatinib (Pemazyre™) EOCCO POLICY
pemigatinib (Pemazyre™) EOCCO POLICY Policy Type: PA/SP Pharmacy Coverage Policy: EOCCO191 Description Pemigatinib (Pemazyre) is an orally administered fibroblast growth factor receptor 2 (FGFR2) inhibitor, with activity against FGFR2 fusions or rearrangements in cholangiocarcinoma cells. Length of Authorization N/A Quantity Limits Product Name Dosage Form Indication Quantity Limit 13.5 mg tablet Previously treated, unresectable, locally Pemigatinib advanced or metastatic 9 mg tablet 14 tablets/21 days (Pemazyre) cholangiocarcinoma in 4.5 mg tablet adults with FGFR2 fusions or rearrangements Initial Evaluation I. Pemigatinib (Pemazyre) is considered investigational when used for all conditions, including but not limited to cholangiocarcinoma. Renewal Evaluation I. N/A Supporting Evidence I. Pemigatinib (Pemazyre) is the first targeted therapy for cholangiocarcinoma that harbors FGFR2 fusions or rearrangements. Pemigatinib (Pemazyre) is a second-line chemotherapy option. Guideline preferred first line chemotherapy is gemcitabine and cisplatin, while second-line options include mFOLFOX, FOLFIRI, and regorafenib (Stivarga). II. Pemigatinib (Pemazyre) was evaluated in FIGHT-202, an open-label, single-arm, multi-cohort Phase 2 trial. Patients (N=146) with locally advanced or metastatic CCA, previously treated with at least 1 chemotherapy were included. FDA approval was based on the overall response rate (ORR) in patients with FGFR2 gene fusion or rearrangements. 1 pemigatinib (Pemazyre™) EOCCO POLICY III. The primary efficacy endpoint was objective response rate (ORR). Secondary endpoints were progression-free survival (PFS), overall survival (OS), and duration of response (DOR). Based on analysis of this clinical trial data, quality of the evidence is considered low given the lack of comparator and open-label trial design, as well as, the lack of clinically meaningful outcomes in morbidity, mortality, and quality of life – medication efficacy has not yet been confirmed. -

PCORI High Potential Disruption Report, November 2020
- PCORI Health Care Horizon Scanning System Volume 2 Issue 2 High Potential Disruption Report November 2020 Prepared for: Patient-Centered Outcomes Research Institute 1828 L St, NW, Suite 900 Washington, DC 20036 Contract No. MSA-HORIZSCAN-ECRI-ENG-2018.7.12 Prepared by: ECRI 5200 Butler Pike Plymouth Meeting, PA 19462 Investigators: Randy Hulshizer, MA, MS Marcus Lynch, PhD, MBA Jennifer De Lurio, MS Damian Carlson, MS Christian Cuevas, PhD Andrea Druga, MSPAS, PA-C Misha Mehta, MS Prital Patel, MPH Brian Wilkinson, MA Donna Beales, MLIS Eloise DeHaan, BS Eileen Erinoff, MSLIS Cassia Hulshizer, AS Madison Kimball, MS Maria Middleton, MPH Diane Robertson, BA Kelley Tipton, MPH Rosemary Walker, MLIS Andrew Furman, MD, MMM, FACEP Statement of Funding and Purpose This report incorporates data collected during implementation of the Patient-Centered Outcomes Research Institute (PCORI) Health Care Horizon Scanning System, operated by ECRI under contract to PCORI, Washington, DC (Contract No. MSA-HORIZSCAN-ECRI-ENG-2018.7.12). The findings and conclusions in this document are those of the authors, who are responsible for its content. No statement in this report should be construed as an official position of PCORI. An intervention that potentially meets inclusion criteria might not appear in this report simply because the horizon scanning system has not yet detected it or it does not yet meet inclusion criteria outlined in the PCORI Health Care Horizon Scanning System: Horizon Scanning Protocol and Operations Manual. Inclusion or absence of interventions in the horizon scanning reports will change over time as new information is collected; therefore, inclusion or absence should not be construed as either an endorsement or rejection of specific interventions.