Potential Exploration of Recent FDA-Approved Anticancer Drugs Against Models of SARS-Cov-2’S Main Protease and Spike Glycoprotein: a Computational Study
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cogent Biosciences Announces Creation of Cogent Research Team
Cogent Biosciences Announces Creation of Cogent Research Team April 6, 2021 Names industry veteran John Robinson, PhD as Chief Scientific Officer New Boulder-based team with exceptional track record of drug discovery and development focused on creating novel small molecule therapies for rare, genetically driven diseases Strong year-end cash position of $242.2 million supports company goals into 2024, including three CGT9486 clinical trials on-track to start this year, beginning with ASM in 1H21 BOULDER, Colo. and CAMBRIDGE, Mass., April 6, 2021 /PRNewswire/ -- Cogent Biosciences, Inc. (Nasdaq: COGT), a biotechnology company focused on developing precision therapies for genetically defined diseases, today announced the formation of the Cogent Research Team led by newly appointed Chief Scientific Officer, John Robinson, PhD. "Today marks an important step forward for Cogent Biosciences as we announce the formation of the Cogent Research Team with a focus on discovering and developing new small molecule therapies for patients fighting rare, genetically driven diseases," said Andrew Robbins, President and Chief Executive Officer of Cogent Biosciences. "I am thrilled to welcome John onboard as Cogent Biosciences' Chief Scientific Officer. John's expertise and seasoned leadership make him ideally suited to lead this new team of world class scientists. Given the team's impressive experience and accomplishments, we are excited for Cogent Biosciences' future and the opportunity to expand our pipeline and deliver novel precision therapies for patients." With an exceptional track record of innovation, the Cogent Research Team will focus on pioneering best-in-class, small molecule therapeutics to both improve upon existing drugs with clear limitations, as well as create new breakthroughs for diseases where others have been unable to find solutions. -

Prior Authorization Oncology – Tukysa™ (Tucatinib Tablets)
Cigna National Formulary Coverage Policy Prior Authorization Oncology – Tukysa™ (tucatinib tablets) Table of Contents Product Identifier(s) National Formulary Medical Necessity ................ 1 64724 Conditions Not Covered....................................... 2 Background .......................................................... 2 References .......................................................... 2 Revision History ................................................... 2 INSTRUCTIONS FOR USE The following Coverage Policy applies to health benefit plans administered by Cigna Companies. Certain Cigna Companies and/or lines of business only provide utilization review services to clients and do not make coverage determinations. References to standard benefit plan language and coverage determinations do not apply to those clients. Coverage Policies are intended to provide guidance in interpreting certain standard benefit plans administered by Cigna Companies. Please note, the terms of a customer’s particular benefit plan document [Group Service Agreement, Evidence of Coverage, Certificate of Coverage, Summary Plan Description (SPD) or similar plan document] may differ significantly from the standard benefit plans upon which these Coverage Policies are based. For example, a customer’s benefit plan document may contain a specific exclusion related to a topic addressed in a Coverage Policy. In the event of a conflict, a customer’s benefit plan document always supersedes the information in the Coverage Policies. In the absence of a controlling federal or state coverage mandate, benefits are ultimately determined by the terms of the applicable benefit plan document. Coverage determinations in each specific instance require consideration of 1) the terms of the applicable benefit plan document in effect on the date of service; 2) any applicable laws/regulations; 3) any relevant collateral source materials including Coverage Policies and; 4) the specific facts of the particular situation. -

Tabrecta (Capmatinib)
Tabrecta (capmatinib) NEW PRODUCT SLIDESHOW Introduction . Brand name: Tabrecta . Generic name: Capmatinib . Pharmacologic class: Kinase inhibitor . Strength and Formulation: 150mg, 200mg; tablets . Manufacturer: Novartis . How supplied: Tabs—56 . Legal Classification: Rx Tabrecta Indication Treatment of adult patients with metastatic non-small cell lung cancer (NSCLC) whose tumors have a mutation that leads to mesenchymal-epithelial transition (MET) exon 14 skipping as detected by an FDA- approved test . Continued approval for this indication may be contingent upon verification and description of clinical benefit in confirmatory trial(s) Dosage and Administration Confirm presence of a mutation that leads to MET exon 14 skipping in tumor specimens Swallow whole 400mg twice daily Recommended dose reductions for adverse reactions: . First: 300mg twice daily . Second: 200mg twice daily . Permanently discontinue in patients unable to tolerate 200mg twice daily Dosage Modifications for Adverse Reactions Interstitial lung disease/pneumonitis . Any grade: permanently discontinue Increased ALT and/or AST without increased total bilirubin . Grade 3: withhold until recovery to baseline ALT/AST; if recovery to baseline within 7 days, then resume at same dose, otherwise resume at a reduced dose . Grade 4: permanently discontinue Increased ALT and/or AST with increased total bilirubin in the absence of cholestasis or hemolysis . ALT and/or AST >3xULN with total bilirubin >2xULN: permanently discontinue Dosage Modifications for Adverse Reactions Increased total bilirubin without concurrent increased ALT and/or AST . Grade 2: withhold until recovery to baseline bilirubin; if recovered to baseline within 7 days, then resume at same dose, otherwise resume at reduced dose . Grade 3: withhold until recovery to baseline bilirubin; if recovered to baseline within 7 days, then resume at reduced dose, otherwise permanently discontinue . -

Incyte Corporation
Building Value through Innovative Medicines 2019 First Quarter Financial and Corporate Update April 30, 2019 Forward-looking Statements Except for the historical information set forth herein, the matters set forth in this presentation contain predictions, estimates and other forward-looking statements, including without limitation statements regarding: expectations regarding ruxolitinib, ruxolitinib cream, itacitinib, pemigatinib, parsaclisib and INCMGA0012 trial results and timing of the receipt and presentation of those results; the expected timing of the NDA submissions for pemigatinib and capmatinib; our belief that certain of our projects, such as the acceleration of vitiligo development and opportunities with pemigatinib and itacitinib, warrant increased near-term funding; expectations regarding planned regulatory updates, planned pivotal clinical updates and planned clinical trial initiations; expectations by our collaborative partners regarding timing of NDA submission for capmatinib and announcement of baricitinib trial results; our plans and expectations for development of, and clinical trials involving, our other product candidates, including the potential timing for regulatory submissions; our plans for immediate launch of ruxolitinib for steroid-refractory acute GVHD should the FDA approve our sNDA; our updated 2019 GAAP and non-GAAP guidance; our expectations regarding baricitinib royalties; and our expected 2019 newsflow events. These forward-looking statements are based on our current expectations and are subject to risks -

Leukemia Insights June 2021
JUNE 2021 In this month’s Leukemia Insights newsletter, written by Prithviraj Bose, M.D. and Srdan Verstovsek, M.D., Ph.D., and sponsored in part by the Charif Souki Cancer Research ABOUT MyMDAnderson Fund, we discuss our novel therapeutic approaches for the rare hematologic malignancies, systemic mastocytosis and myeloid/lymphoid neoplasms with myMDAnderson is a secure, eosinophilia. Learn more about our Leukemia program. personalized web site helping community physicians expedite patient referrals, as well as improve continuity Spotlight on rare, atypical, myeloid of care through information access and streamlined communications. neoplasms: systemic mastocytosis and Physicians who have referred patients myeloid/lymphoid neoplasms with to MD Anderson or plan to do so, can utilize the HIPAA compliant features eosinophilia and FGFR1 rearrangements of myMDAnderson to: • Refer a patient • View your patient's appointments Systemic Mastocytosis Access patient reports • Send and receive secure Systemic mastocytosis (SM) is a rare myeloid neoplasm messages driven in approximately 95% of cases by an activating mutation in c-KIT, usually D816V. SM is characterized as indolent, smoldering or advanced, based on the presence JOIN THE COVERSATION and number of so-called B- and C-findings. The latter Connect with us. signify organ damage and are a hallmark of advanced SM (AdvSM). AdvSM, in turn, is typically sub-classified as aggressive SM (ASM), SM with an associated hematologic neoplasm (SM-AHN, the most common subtype) and mast cell leukemia (MCL). Patients with indolent SM (ISM) and smoldering SM (SSM) enjoy much better survival than JOIN OUR MAILING LIST those with AdvSM, although symptoms in all three subtypes can be severe and debilitating. -

Incyte Announces the European Commission Approval of Pemazyre
3/29/2021 Incyte Announces the European Commission Approval of Pemazyre® (pemigatinib) as a Treatment for Adults with Locally Advanced or Metastatic Cholangiocarcinoma with a Fibroblast Growth Factor Receptor 2 (FGFR2) Fusion or Rearrangement - Pemazyre is the rst targeted therapy approved in the EU for this indication WILMINGTON, Del.--(BUSINESS WIRE)-- Incyte (Nasdaq:INCY) today announced that the European Commission (EC) has approved Pemazyre® (pemigatinib) for the treatment of adults with locally advanced or metastatic cholangiocarcinoma with a broblast growth factor receptor 2 (FGFR2) fusion or rearrangement that have progressed after at least one prior line of systemic therapy. The decision follows the positive opinion received from the European Medicines Agency’s Committee for Medicinal Products for Human Use in January 2021 recommending the conditional marketing authorization of Pemazyre. “Pemazyre’s approval is a crucial milestone for patients with FGFR2 positive cholangiocarcinoma. It is the rst new treatment option to be made available to these patients in the EU in over a decade and has demonstrated a high rate of durable responses in a setting where historically there has been no eective standard of care,” said Hervé Hoppenot, Chief Executive Ocer, Incyte. “We now look forward to working with individual countries in Europe to ensure eligible patients can access this new treatment as soon as possible.” The EC decision is based on data from the FIGHT-202 study evaluating the safety and ecacy of Pemazyre in adult patients with previously treated, locally advanced or metastatic cholangiocarcinoma with documented FGF/FGFR status. Interim results from FIGHT-202 demonstrated that in patients harboring FGFR2 fusions or rearrangements (Cohort A [108 patients]), Pemazyre monotherapy resulted in an overall response rate (ORR) of 37 percent (primary endpoint) and a median duration of response (DOR) of 8 months (secondary endpoint) based on an independent central radiographic review. -

Insights from Drug Discovery Life Cycle Management in Pharmaceutical Industry: a Case-Study B
et International Journal on Emerging Technologies 10 (4): 212-217(2019) ISSN No. (Print): 0975-8364 ISSN No. (Online): 2249-3255 Insights from Drug Discovery Life Cycle Management in Pharmaceutical Industry: A Case-study B. Thomas 1 and P. Chugan 2 1Research Scholar, Institute of Management, Nirma University, Ahmedabad (Gujarat), India. 2Professor (Retd.), Institute of Management, Nirma University, Ahmedabad (Gujarat), India. (Corresponding author: B. Thomas) (Received 20 September 2019, Revised 16 November 2019, Accepted 22 November 2019) (Published by Research Trend, Website: www.researchtrend.net) ABSTRACT: The Biotechnology and Pharmaceutical Industry has always stood up to the challenge of discovering innovative products fulfilling high unmet medical needs for mankind. The drug discovery and development pathway is very challenging and time consuming from efficacy, safety, regulatory and investments perspective. Hence, there is a significant need for collaboration among the biotechnology and large pharmaceutical companies to develop and commercialize their product innovations across the globe. This paper reviews the discovery and development of Selumetinib by Array Biopharma from year 2000 to 2019 when the USFDA accepted the first NDA filing for neurofibromatosis type one (NF1) disease. Array biopharma successfully developed several other drugs to the marketplace and several drugs to late stage registration trials either itself or in collaboration with strategic partners. These strategic partnerships helped mitigate risks in drug development and registration. This study of the clinical, regulatory and partnership strategy untaken for Selumetinib has created important contribution in the field of product life cycle management in the pharmaceutical and biotechnology industry. Keywords: Product development, drug development, life-cycle management, strategic partnership. -

Clinicogenomic Analysis of FGFR2-Rearranged Cholangiocarcinoma Identifies Correlates of Response and Mechanisms of Resistance to Pemigatinib
Published OnlineFirst November 20, 2020; DOI: 10.1158/2159-8290.CD-20-0766 RESEARCH ARTICLE Clinicogenomic Analysis of FGFR2- Rearranged Cholangiocarcinoma Identifies Correlates of Response and Mechanisms of Resistance to Pemigatinib Ian M. Silverman1, Antoine Hollebecque2, Luc Friboulet2, Sherry Owens1, Robert C. Newton1, Huiling Zhen3, Luis Féliz4, Camilla Zecchetto5, Davide Melisi5, and Timothy C. Burn1 Downloaded from cancerdiscovery.aacrjournals.org on September 30, 2021. © 2020 American Association for Cancer Research. Published OnlineFirst November 20, 2020; DOI: 10.1158/2159-8290.CD-20-0766 ABSTRACT Pemigatinib, a selective FGFR1–3 inhibitor, has demonstrated antitumor activity in FIGHT-202, a phase II study in patients with cholangiocarcinoma harboring FGFR2 fusions/rearrangements, and has gained regulatory approval in the United States. Eligibility for FIGHT- 202 was assessed using genomic profiling; here, these data were utilized to characterize the genomic landscape of cholangiocarcinoma and to uncover unique molecular features of patients harboring FGFR2 rearrangements. The results highlight the high percentage of patients with cholangiocarcinoma harboring potentially actionable genomic alterations and the diversity in gene partners that rear- range with FGFR2. Clinicogenomic analysis of pemigatinib-treated patients identified mechanisms of primary and acquired resistance. Genomic subsets of patients with other potentially actionable FGF/ FGFR alterations were also identified. Our study provides a framework for molecularly guided clinical trials and underscores the importance of genomic profiling to enable a deeper understanding of the molecular basis for response and nonresponse to targeted therapy. SIGNIFICANCE: We utilized genomic profiling data from FIGHT-202 to gain insights into the genomic landscape of cholangiocarcinoma, to understand the molecular diversity of patients with FGFR2 fusions or rearrangements, and to interrogate the clinicogenomics of patients treated with pemi- gatinib. -

First-Line Treatment Options for Patients with Stage IV Non-Small Cell Lung Cancer with Driver Alterations
First-Line Treatment Options for Patients with Stage IV Non-Small Cell Lung Cancer with Driver Alterations Patients with stage IV non-small cell lung cancer Nonsquamous cell carcinoma and squamous cell carcinoma Activating EGFR mutation other Sensitizing (L858R/exon 19 MET exon 14 skipping than exon 20 insertion mutations, EGFR exon 20 mutation ALK rearrangement ROS1 rearrangement BRAF V600E mutation RET rearrangement NTRK rearrangement mutations KRAS alterations HER2 alterations NRG1 alterations deletion) EGFR mutation T790M, L858R or Ex19Del PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options Emerging target; no Emerging target; no Emerging target; no Platinum doublet † † † Osimertinib monotherapy S Afatinib monotherapy M M Alectinib S Entrectinib M Dabrafenib/trametinib M Capmatinib M Selpercatinib M Entrectinib M conclusions available conclusions available conclusions available chemotherapy ± bevacizumab Gefitinib with doublet Standard treatment based on Standard treatment based on Standard treatment based on M M M Brigatinib S Crizotinib M M Tepotinib M Pralsetinib* W Larotrectinib M chemotherapy non-driver mutation guideline non-driver mutation guideline non-driver mutation guideline If alectinib or brigatinib are not available If entrectinib or crizotinib are not available Standard treatment based on Standard treatment based on Standard treatment based on Dacomitinib monotherapy M Osimertinib W M M M Ceritinib S Ceritinib W non-driver mutation guideline non-driver mutation guideline non-driver mutation guideline Monotherapy with afatinib M Crizotinib S Lortlatinib W Standard treatment based on Erlotinib/ramucirumab M M non-driver mutation guideline Erlotinib/bevacizumab M Monotherapy with erlotinib M Strength of Recommendation Monotherapy with gefitinib M S Strong M Moderate W Weak Monotherapy with icotinib M Notes. -
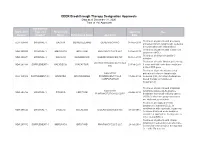
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities -

Overcoming MET-Dependent Resistance to Selective RET Inhibition in Patients with RET Fusion–Positive Lung Cancer by Combining Selpercatinib with Crizotinib a C Ezra Y
Published OnlineFirst October 20, 2020; DOI: 10.1158/1078-0432.CCR-20-2278 CLINICAL CANCER RESEARCH | RESEARCH BRIEFS: CLINICAL TRIAL BRIEF REPORT Overcoming MET-Dependent Resistance to Selective RET Inhibition in Patients with RET Fusion–Positive Lung Cancer by Combining Selpercatinib with Crizotinib A C Ezra Y. Rosen1, Melissa L. Johnson2, Sarah E. Clifford3, Romel Somwar4, Jennifer F. Kherani5, Jieun Son3, Arrien A. Bertram3, Monika A. Davare6, Eric Gladstone4, Elena V. Ivanova7, Dahlia N. Henry5, Elaine M. Kelley3, Mika Lin3, Marina S.D. Milan3, Binoj C. Nair5, Elizabeth A. Olek5, Jenna E. Scanlon3, Morana Vojnic4, Kevin Ebata5, Jaclyn F. Hechtman4, Bob T. Li1,8, Lynette M. Sholl9, Barry S. Taylor10, Marc Ladanyi4, Pasi A. Janne€ 3, S. Michael Rothenberg5, Alexander Drilon1,8, and Geoffrey R. Oxnard3 ABSTRACT ◥ Purpose: The RET proto-oncogene encodes a receptor tyrosine Results: MET amplification was identified in posttreatment kinase that is activated by gene fusion in 1%–2% of non–small biopsies in 4 patients with RET fusion–positive NSCLC treated with cell lung cancers (NSCLC) and rarely in other cancer types. selpercatinib. In at least one case, MET amplification was clearly Selpercatinib is a highly selective RET kinase inhibitor that has evident prior to therapy with selpercatinib. We demonstrate that recently been approved by the FDA in lung and thyroid cancers increased MET expression in RET fusion–positive tumor cells causes with activating RET gene fusions and mutations. Molecular resistance to selpercatinib, and this can be overcome by combining mechanisms of acquired resistance to selpercatinib are poorly selpercatinib with crizotinib. Using SPPs, selpercatinib with crizo- understood. -

Rxoutlook® 1St Quarter 2019
® RxOutlook 1st Quarter 2020 optum.com/optumrx a RxOutlook 1st Quarter 2020 Orphan drugs continue to feature prominently in the drug development pipeline In 1983 the Orphan Drug Act was signed into law. Thirty seven years later, what was initially envisioned as a minor category of drugs has become a major part of the drug development pipeline. The Orphan Drug Act was passed by the United States Congress in 1983 in order to spur drug development for rare conditions with high unmet need. The legislation provided financial incentives to manufacturers if they could demonstrate that the target population for their drug consisted of fewer than 200,000 persons in the United States, or that there was no reasonable expectation that commercial sales would be sufficient to recoup the developmental costs associated with the drug. These “Orphan Drug” approvals have become increasingly common over the last two decades. In 2000, two of the 27 (7%) new drugs approved by the FDA had Orphan Designation, whereas in 2019, 20 of the 48 new drugs (42%) approved by the FDA had Orphan Designation. Since the passage of the Orphan Drug Act, 37 years ago, additional regulations and FDA designations have been implemented in an attempt to further expedite drug development for certain serious and life threatening conditions. Drugs with a Fast Track designation can use Phase 2 clinical trials to support FDA approval. Drugs with Breakthrough Therapy designation can use alternative clinical trial designs instead of the traditional randomized, double-blind, placebo-controlled trial. Additionally, drugs may be approved via the Accelerated Approval pathway using surrogate endpoints in clinical trials rather than clinical outcomes.