RET Inhibition in Novel Patient-Derived Models of RET Fusion
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cstone's Partner Blueprint Medicines Presented Updated Part 1 Data from PIONEER Trial of Avapritinib
Hong Kong Exchanges and Clearing Limited and The Stock Exchange of Hong Kong Limited take no responsibility for the contents of this announcement, make no representation as to its accuracy or completeness and expressly disclaim any liability whatsoever for any loss howsoever arising from or in reliance upon the whole or any part of the contents of this announcement. The forward-looking statements made in this announcement relate only to the events or information as of the date on which the statements are made in this announcement. Except as required by law, we undertake no obligation to update or revise publicly any forward-looking statements, whether as a result of new information, future events or otherwise, after the date on which the statements are made or to reflect the occurrence of unanticipated events. You should read this announcement completely and with the understanding that our actual future results or performance may be materially different from what we expect. In this announcement, statements of, or references to, our intentions or those of any of our directors and/or our Company are made as of the date of this announcement. Any of these intentions may alter in light of future development. CStone Pharmaceuticals 基 石 藥 業 (Incorporated in the Cayman Islands with limited liability) (Stock Code: 2616) VOLUNTARY ANNOUNCEMENT CSTONE’S PARTNER BLUEPRINT MEDICINES PRESENTED UPDATED PART 1 DATA FROM PIONEER TRIAL OF AVAPRITINIB SHOWING ROBUST REDUCTIONS IN CUTANEOUS DISEASE SYMPTOMS IN PATIENTS WITH INDOLENT SYSTEMIC MASTOCYTOSIS The partner of CStone Pharmaceuticals (the “Company” or “CStone”), Blueprint Medicines Corporation (NASDAQ: BPMC) (“Blueprint Medicines”), announced on June 6 updated clinical data from part 1 of the PIONEER trial showing robust and consistent clinical activity for avapritinib across multiple qualitative and quantitative measures of cutaneous disease in patients with indolent systemic mastocytosis (“ISM”). -

Functional Roles of Bromodomain Proteins in Cancer
cancers Review Functional Roles of Bromodomain Proteins in Cancer Samuel P. Boyson 1,2, Cong Gao 3, Kathleen Quinn 2,3, Joseph Boyd 3, Hana Paculova 3 , Seth Frietze 3,4,* and Karen C. Glass 1,2,4,* 1 Department of Pharmaceutical Sciences, Albany College of Pharmacy and Health Sciences, Colchester, VT 05446, USA; [email protected] 2 Department of Pharmacology, Larner College of Medicine, University of Vermont, Burlington, VT 05405, USA; [email protected] 3 Department of Biomedical and Health Sciences, University of Vermont, Burlington, VT 05405, USA; [email protected] (C.G.); [email protected] (J.B.); [email protected] (H.P.) 4 University of Vermont Cancer Center, Burlington, VT 05405, USA * Correspondence: [email protected] (S.F.); [email protected] (K.C.G.) Simple Summary: This review provides an in depth analysis of the role of bromodomain-containing proteins in cancer development. As readers of acetylated lysine on nucleosomal histones, bromod- omain proteins are poised to activate gene expression, and often promote cancer progression. We examined changes in gene expression patterns that are observed in bromodomain-containing proteins and associated with specific cancer types. We also mapped the protein–protein interaction network for the human bromodomain-containing proteins, discuss the cellular roles of these epigenetic regu- lators as part of nine different functional groups, and identify bromodomain-specific mechanisms in cancer development. Lastly, we summarize emerging strategies to target bromodomain proteins in cancer therapy, including those that may be essential for overcoming resistance. Overall, this review provides a timely discussion of the different mechanisms of bromodomain-containing pro- Citation: Boyson, S.P.; Gao, C.; teins in cancer, and an updated assessment of their utility as a therapeutic target for a variety of Quinn, K.; Boyd, J.; Paculova, H.; cancer subtypes. -

Trim33 Is Essential for Macrophage and Neutrophil Mobilization To
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 2797-2807 doi:10.1242/jcs.203471 RESEARCH ARTICLE Trim33 is essential for macrophage and neutrophil mobilization to developmental or inflammatory cues Doris Lou Demy1,2,*, Muriel Tauzin1,2,‡, Mylenè Lancino1,2,Véronique Le Cabec3,4, Michael Redd5, Emi Murayama1,2, Isabelle Maridonneau-Parini3,4, Nikolaus Trede5 and Philippe Herbomel1,2,§ ABSTRACT first organs to become colonized in this way in mammalian and Macrophages infiltrate and establish in developing organs from an avian embryogenesis is the central nervous system (CNS) (Sorokin early stage, often before these have become vascularized. Similarly, et al., 1992; Cuadros et al., 1993), and at least in mammals, the leukocytes, in general, can quickly migrate through tissues to any site resulting resident macrophages then comprise the microglia for the of wounding. This unique capacity is rooted in their characteristic whole life of the animal (Kierdorf et al., 2015). The molecular and amoeboid motility, the genetic basis of which is poorly understood. cellular determinants of this early colonization are mostly unknown. Trim33 (also known as Tif1-γ), a nuclear protein that associates In the zebrafish embryo, primitive macrophages born in the yolk sac with specific DNA-binding transcription factors to modulate gene colonize the tissues of the embryo, notably the head, brain and expression, has been found to be mainly involved in hematopoiesis retina, in a pattern very similar to that seen in mammalian and avian and gene regulation mediated by TGF-β. Here, we have discovered embryos (Herbomel et al., 2001). The M-CSF/CSF-1 receptor that in Trim33-deficient zebrafish embryos, primitive macrophages are (CSF1-R) was found to be dispensable for the differentiation of unable to colonize the central nervous system to become microglia. -

Clinical and Molecular Relevance of Genetic Variants in the Non-Coding
Clinical and molecular relevance of genetic variants in the non-coding transcriptome of patients with cytogenetically normal acute myeloid leukemia by Dimitrios Papaioannou, Hatice G. Ozer, Deedra Nicolet, Amog P. Urs, Tobias Herold, Krzysztof Mrózek, Aarif M.N. Batcha, Klaus H. Metzeler, Ayse S. Yilmaz, Stefano Volinia, Marius Bill, Jessica Kohlschmidt, Maciej Pietrzak, Christopher J. Walker, Andrew J. Carroll, Jan Braess, Bayard L. Powell, Ann-Kathrin Eisfeld, Geoffrey L. Uy, Eunice S. Wang, Jonathan E. Kolitz, Richard M. Stone, Wolfgang Hiddemann, John Byrd, Clara D. Bloomfield, and Ramiro Garzon Haematologica 2021 [Epub ahead of print] Citation: Dimitrios Papaioannou, Hatice G. Ozer, Deedra Nicolet, Amog P. Urs, Tobias Herold, Krzysztof Mrózek, Aarif M.N. Batcha, Klaus H. Metzeler, Ayse S. Yilmaz, Stefano Volinia, Marius Bill, Jessica Kohlschmidt, Maciej Pietrzak, Christopher J. Walker, Andrew J. Carroll, Jan Braess, Bayard L. Powell, Ann-Kathrin Eisfeld, Geoffrey L. Uy, Eunice S. Wang, Jonathan E. Kolitz, Richard M. Stone, Wolfgang Hiddemann, John Byrd, Clara D. Bloomfield, and Ramiro Garzon. Clinical and molecular relevance of genetic variants in the non-coding transcriptome of patients with cytogenetically normal acute myeloid leukemia. Haematologica. 2021; 106:xxx doi:10.3324/haematol.2021.266643 Publisher's Disclaimer. E-publishing ahead of print is increasingly important for the rapid dissemination of science. Haematologica is, therefore, E-publishing PDF files of an early version of manuscripts that have completed a regular peer review and have been accepted for publication. E-publishing of this PDF file has been approved by the authors. After having E-published Ahead of Print, manuscripts will then undergo technical and English editing, typesetting, proof correction and be presented for the authors' final approval; the final version of the manuscript will then appear in print on a regular issue of the journal. -

Role of CREB/CRTC1-Regulated Gene Transcription During Hippocampal-Dependent Memory in Alzheimer’S Disease Mouse Models
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Institut de Neurociències Universitat Autònoma de Barcelona Departament de Bioquímica i Biologia Molecular Unitat de Bioquímica, Facultat de Medicina Role of CREB/CRTC1-regulated gene transcription during hippocampal-dependent memory in Alzheimer’s disease mouse models Arnaldo J. Parra Damas TESIS DOCTORAL Bellaterra, 2015 Institut de Neurociències Departament de Bioquímica i Biologia Molecular Universitat Autònoma de Barcelona Role of CREB/CRTC1-regulated gene transcription during hippocampal-dependent memory in Alzheimer’s disease mouse models Papel de la transcripción génica regulada por CRTC1/CREB durante memoria dependiente de hipocampo en modelos murinos de la enfermedad de Alzheimer Memoria de tesis doctoral presentada por Arnaldo J. Parra Damas para optar al grado de Doctor en Neurociencias por la Universitat Autonòma de Barcelona. Trabajo realizado en la Unidad de Bioquímica y Biología Molecular de la Facultad de Medicina del Departamento de Bioquímica y Biología Molecular de la Universitat Autònoma de Barcelona, y en el Instituto de Neurociencias de la Universitat Autònoma de Barcelona, bajo la dirección del Doctor Carlos Saura Antolín. -

Cogent Biosciences Announces Creation of Cogent Research Team
Cogent Biosciences Announces Creation of Cogent Research Team April 6, 2021 Names industry veteran John Robinson, PhD as Chief Scientific Officer New Boulder-based team with exceptional track record of drug discovery and development focused on creating novel small molecule therapies for rare, genetically driven diseases Strong year-end cash position of $242.2 million supports company goals into 2024, including three CGT9486 clinical trials on-track to start this year, beginning with ASM in 1H21 BOULDER, Colo. and CAMBRIDGE, Mass., April 6, 2021 /PRNewswire/ -- Cogent Biosciences, Inc. (Nasdaq: COGT), a biotechnology company focused on developing precision therapies for genetically defined diseases, today announced the formation of the Cogent Research Team led by newly appointed Chief Scientific Officer, John Robinson, PhD. "Today marks an important step forward for Cogent Biosciences as we announce the formation of the Cogent Research Team with a focus on discovering and developing new small molecule therapies for patients fighting rare, genetically driven diseases," said Andrew Robbins, President and Chief Executive Officer of Cogent Biosciences. "I am thrilled to welcome John onboard as Cogent Biosciences' Chief Scientific Officer. John's expertise and seasoned leadership make him ideally suited to lead this new team of world class scientists. Given the team's impressive experience and accomplishments, we are excited for Cogent Biosciences' future and the opportunity to expand our pipeline and deliver novel precision therapies for patients." With an exceptional track record of innovation, the Cogent Research Team will focus on pioneering best-in-class, small molecule therapeutics to both improve upon existing drugs with clear limitations, as well as create new breakthroughs for diseases where others have been unable to find solutions. -

The Essential Role of the Crtc2-CREB Pathway in Β Cell Function And
The Essential Role of the Crtc2-CREB Pathway in β cell Function and Survival by Chandra Eberhard A thesis submitted to the School of Graduate Studies and Research in partial fulfillment of the requirements for the degree of Masters of Science in Cellular and Molecular Medicine Department of Cellular and Molecular Medicine Faculty of Medicine University of Ottawa September 28, 2012 © Chandra Eberhard, Ottawa, Canada, 2013 Abstract: Immunosuppressants that target the serine/threonine phosphatase calcineurin are commonly administered following organ transplantation. Their chronic use is associated with reduced insulin secretion and new onset diabetes in a subset of patients, suggestive of pancreatic β cell dysfunction. Calcineurin plays a critical role in the activation of CREB, a key transcription factor required for β cell function and survival. CREB activity in the islet is activated by glucose and cAMP, in large part due to activation of Crtc2, a critical coactivator for CREB. Previous studies have demonstrated that Crtc2 activation is dependent on dephosphorylation regulated by calcineurin. In this study, we sought to evaluate the impact of calcineurin-inhibiting immunosuppressants on Crtc2-CREB activation in the primary β cell. In addition, we further characterized the role and regulation of Crtc2 in the β cell. We demonstrate that Crtc2 is required for glucose dependent up-regulation of CREB target genes. The phosphatase calcineurin and kinase regulation by LKB1 contribute to the phosphorylation status of Crtc2 in the β cell. CsA and FK506 block glucose-dependent dephosphorylation and nuclear translocation of Crtc2. Overexpression of a constitutively active mutant of Crtc2 that cannot be phosphorylated at Ser171 and Ser275 enables CREB activity under conditions of calcineurin inhibition. -

Trim24 Targets Endogenous P53 for Degradation
Trim24 targets endogenous p53 for degradation Kendra Alltona,b,1, Abhinav K. Jaina,b,1, Hans-Martin Herza, Wen-Wei Tsaia,b, Sung Yun Jungc, Jun Qinc, Andreas Bergmanna, Randy L. Johnsona,b, and Michelle Craig Bartona,b,2 aDepartment of Biochemistry and Molecular Biology, Program in Genes and Development, Graduate School of Biomedical Sciences and bCenter for Stem Cell and Developmental Biology, University of Texas M.D. Anderson Cancer Center, 1515 Holcombe Boulevard, Houston, TX 77030; and cDepartment of Molecular and Cellular Biology, Baylor College of Medicine, 1 Baylor Plaza, Houston, TX 77030 Edited by Carol L. Prives, Columbia University, New York, NY, and approved May 15, 2009 (received for review December 23, 2008) Numerous studies focus on the tumor suppressor p53 as a protector regulator of p53 suggests that potential therapeutic targets to of genomic stability, mediator of cell cycle arrest and apoptosis, restore p53 functions remain to be identified. and target of mutation in 50% of all human cancers. The vast majority of information on p53, its protein-interaction partners and Results and Discussion regulation, comes from studies of tumor-derived, cultured cells Creation of a Mouse and Stem Cell Model for p53 Analysis. To where p53 and its regulatory controls may be mutated or dysfunc- facilitate analysis of endogenous p53 in normal cells, we per- tional. To address regulation of endogenous p53 in normal cells, formed gene targeting to create mouse embryonic stem (ES) we created a mouse and stem cell model by knock-in (KI) of a cells and mice (12), which express endogenously regulated p53 tandem-affinity-purification (TAP) epitope at the endogenous protein fused with a C-terminal TAP tag (p53-TAPKI, Fig. -

Transcriptomic and Proteomic Profiling Provides Insight Into
BASIC RESEARCH www.jasn.org Transcriptomic and Proteomic Profiling Provides Insight into Mesangial Cell Function in IgA Nephropathy † † ‡ Peidi Liu,* Emelie Lassén,* Viji Nair, Celine C. Berthier, Miyuki Suguro, Carina Sihlbom,§ † | † Matthias Kretzler, Christer Betsholtz, ¶ Börje Haraldsson,* Wenjun Ju, Kerstin Ebefors,* and Jenny Nyström* *Department of Physiology, Institute of Neuroscience and Physiology, §Proteomics Core Facility at University of Gothenburg, University of Gothenburg, Gothenburg, Sweden; †Division of Nephrology, Department of Internal Medicine and Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan; ‡Division of Molecular Medicine, Aichi Cancer Center Research Institute, Nagoya, Japan; |Department of Immunology, Genetics and Pathology, Uppsala University, Uppsala, Sweden; and ¶Integrated Cardio Metabolic Centre, Karolinska Institutet Novum, Huddinge, Sweden ABSTRACT IgA nephropathy (IgAN), the most common GN worldwide, is characterized by circulating galactose-deficient IgA (gd-IgA) that forms immune complexes. The immune complexes are deposited in the glomerular mesangium, leading to inflammation and loss of renal function, but the complete pathophysiology of the disease is not understood. Using an integrated global transcriptomic and proteomic profiling approach, we investigated the role of the mesangium in the onset and progression of IgAN. Global gene expression was investigated by microarray analysis of the glomerular compartment of renal biopsy specimens from patients with IgAN (n=19) and controls (n=22). Using curated glomerular cell type–specific genes from the published literature, we found differential expression of a much higher percentage of mesangial cell–positive standard genes than podocyte-positive standard genes in IgAN. Principal coordinate analysis of expression data revealed clear separation of patient and control samples on the basis of mesangial but not podocyte cell–positive standard genes. -

Loss of TRIM33 Causes Resistance to BET Bromodomain Inhibitors Through MYC- and TGF-Β–Dependent Mechanisms
Loss of TRIM33 causes resistance to BET PNAS PLUS bromodomain inhibitors through MYC- and TGF-β–dependent mechanisms Xiarong Shia, Valia T. Mihaylovaa, Leena Kuruvillaa, Fang Chena, Stephen Vivianoa, Massimiliano Baldassarrea, David Sperandiob, Ruben Martinezb, Peng Yueb, Jamie G. Batesb, David G. Breckenridgeb, Joseph Schlessingera,1, Benjamin E. Turka,1, and David A. Calderwooda,c,1 aDepartment of Pharmacology, Yale University School of Medicine, New Haven, CT 06520; bGilead Sciences, Foster City, CA 94404; and cDepartment of Cell Biology, Yale University School of Medicine, New Haven, CT 06520 Contributed by Joseph Schlessinger, May 24, 2016 (sent for review December 22, 2015; reviewed by Gary L. Johnson and Michael B. Yaffe) Bromodomain and extraterminal domain protein inhibitors (BETi) not characterized by genetic alterations in BET proteins. One key hold great promise as a novel class of cancer therapeutics. Because mechanism by which BETi suppress growth and survival of at least acquired resistance typically limits durable responses to targeted some types of cancer cells is by preferentially repressing tran- therapies, it is important to understand mechanisms by which scription of the proto-oncogene MYC, which is often under the tumor cells adapt to BETi. Here, through pooled shRNA screening control of BRD4 (5, 10, 12, 18). Thus, BETi may provide a new of colorectal cancer cells, we identified tripartite motif-containing mechanism to target MYC and other oncogenic transcription fac- protein 33 (TRIM33) as a factor promoting sensitivity to BETi. We tors, which lack obvious binding pockets for small molecules and are demonstrate that loss of TRIM33 reprograms cancer cells to a more thus typically considered to be “undruggable.” resistant state through at least two mechanisms. -

First-Line Treatment Options for Patients with Stage IV Non-Small Cell Lung Cancer with Driver Alterations
First-Line Treatment Options for Patients with Stage IV Non-Small Cell Lung Cancer with Driver Alterations Patients with stage IV non-small cell lung cancer Nonsquamous cell carcinoma and squamous cell carcinoma Activating EGFR mutation other Sensitizing (L858R/exon 19 MET exon 14 skipping than exon 20 insertion mutations, EGFR exon 20 mutation ALK rearrangement ROS1 rearrangement BRAF V600E mutation RET rearrangement NTRK rearrangement mutations KRAS alterations HER2 alterations NRG1 alterations deletion) EGFR mutation T790M, L858R or Ex19Del PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options PS 0-2 Treatment Options Emerging target; no Emerging target; no Emerging target; no Platinum doublet † † † Osimertinib monotherapy S Afatinib monotherapy M M Alectinib S Entrectinib M Dabrafenib/trametinib M Capmatinib M Selpercatinib M Entrectinib M conclusions available conclusions available conclusions available chemotherapy ± bevacizumab Gefitinib with doublet Standard treatment based on Standard treatment based on Standard treatment based on M M M Brigatinib S Crizotinib M M Tepotinib M Pralsetinib* W Larotrectinib M chemotherapy non-driver mutation guideline non-driver mutation guideline non-driver mutation guideline If alectinib or brigatinib are not available If entrectinib or crizotinib are not available Standard treatment based on Standard treatment based on Standard treatment based on Dacomitinib monotherapy M Osimertinib W M M M Ceritinib S Ceritinib W non-driver mutation guideline non-driver mutation guideline non-driver mutation guideline Monotherapy with afatinib M Crizotinib S Lortlatinib W Standard treatment based on Erlotinib/ramucirumab M M non-driver mutation guideline Erlotinib/bevacizumab M Monotherapy with erlotinib M Strength of Recommendation Monotherapy with gefitinib M S Strong M Moderate W Weak Monotherapy with icotinib M Notes. -
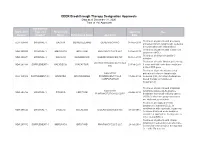
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities