Common Variants in Mendelian Kidney Disease Genes and Their Association with Renal Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
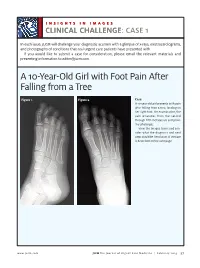
A 10-Year-Old Girl with Foot Pain After Falling from a Tree
INSIGHTS IN IMAGES CLINICAL CHALLENGECHALLENGE: CASE 1 In each issue, JUCM will challenge your diagnostic acumen with a glimpse of x-rays, electrocardiograms, and photographs of conditions that real urgent care patients have presented with. If you would like to submit a case for consideration, please email the relevant materials and presenting information to [email protected]. A 10-Year-Old Girl with Foot Pain After Falling from a Tree Figure 1. Figure 2. Case A 10-year-old girl presents with pain after falling from a tree, landing on her right foot. On examination, the pain emanates from the second through fifth metatarsals and proxi- mal phalanges. View the images taken and con- sider what the diagnosis and next steps would be. Resolution of the case is described on the next page. www.jucm.com JUCM The Journal of Urgent Care Medicine | February 2019 37 INSIGHTS IN IMAGES: CLINICAL CHALLENGE THE RESOLUTION Figure 1. Mediastinal air Figure 1. Differential Diagnosis Pearls for Urgent Care Management and Ⅲ Fracture of the distal fourth metatarsal Considerations for Transfer Ⅲ Plantar plate disruption Ⅲ Emergent transfer should be considered with associated neu- Ⅲ Sesamoiditis rologic deficit, compartment syndrome, open fracture, or vas- Ⅲ Turf toe cular compromise Ⅲ Referral to an orthopedist is warranted in the case of an in- Diagnosis tra-articular fracture, or with Lisfranc ligament injury or ten- Angulation of the distal fourth metatarsal metaphyseal cortex derness over the Lisfranc ligament and hairline lucency consistent with fracture. Acknowledgment: Images courtesy of Teleradiology Associates. Learnings/What to Look for Ⅲ Proximal metatarsal fractures are most often caused by crush- ing or direct blows Ⅲ In athletes, an axial load placed on a plantar-flexed foot should raise suspicion of a Lisfranc injury 38 JUCM The Journal of Urgent Care Medicine | February 2019 www.jucm.com INSIGHTS IN IMAGES CLINICAL CHALLENGE: CASE 2 A 55-Year-Old Man with 3 Hours of Epigastric Pain 55 years PR 249 QRSD 90 QT 471 QTc 425 AXES P 64 QRS -35 T 30 Figure 1. -

Structure and Function of the Human Recq DNA Helicases
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2005 Structure and function of the human RecQ DNA helicases Garcia, P L Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-34420 Dissertation Published Version Originally published at: Garcia, P L. Structure and function of the human RecQ DNA helicases. 2005, University of Zurich, Faculty of Science. Structure and Function of the Human RecQ DNA Helicases Dissertation zur Erlangung der naturwissenschaftlichen Doktorw¨urde (Dr. sc. nat.) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultat¨ der Universitat¨ Z ¨urich von Patrick L. Garcia aus Unterseen BE Promotionskomitee Prof. Dr. Josef Jiricny (Vorsitz) Prof. Dr. Ulrich H ¨ubscher Dr. Pavel Janscak (Leitung der Dissertation) Z ¨urich, 2005 For my parents ii Summary The RecQ DNA helicases are highly conserved from bacteria to man and are required for the maintenance of genomic stability. All unicellular organisms contain a single RecQ helicase, whereas the number of RecQ homologues in higher organisms can vary. Mu- tations in the genes encoding three of the five human members of the RecQ family give rise to autosomal recessive disorders called Bloom syndrome, Werner syndrome and Rothmund-Thomson syndrome. These diseases manifest commonly with genomic in- stability and a high predisposition to cancer. However, the genetic alterations vary as well as the types of tumours in these syndromes. Furthermore, distinct clinical features are observed, like short stature and immunodeficiency in Bloom syndrome patients or premature ageing in Werner Syndrome patients. Also, the biochemical features of the human RecQ-like DNA helicases are diverse, pointing to different roles in the mainte- nance of genomic stability. -

Next Generation Sequencing Panels for Disorders of Sex Development
Next Generation Sequencing Panels for Disorders of Sex Development Disorders of Sex Development – Overview Disorders of sex development (DSDs) occur when sex development does not follow the course of typical male or female patterning. Types of DSDs include congenital development of ambiguous genitalia, disjunction between the internal and external sex anatomy, incomplete development of the sex anatomy, and abnormalities of the development of gonads (such as ovotestes or streak ovaries) (1). Sex chromosome anomalies including Turner syndrome and Klinefelter syndrome as well as sex chromosome mosaicism are also considered to be DSDs. DSDs can be caused by a wide range of genetic abnormalities (2). Determining the etiology of a patient’s DSD can assist in deciding gender assignment, provide recurrence risk information for future pregnancies, and can identify potential health problems such as adrenal crisis or gonadoblastoma (1, 3). Sex chromosome aneuploidy and copy number variation are common genetic causes of DSDs. For this reason, chromosome analysis and/or microarray analysis typically should be the first genetic analysis in the case of a patient with ambiguous genitalia or other suspected disorder of sex development. Identifying whether a patient has a 46,XY or 46,XX karyotype can also be helpful in determining appropriate additional genetic testing. Abnormal/Ambiguous Genitalia Panel Our Abnormal/Ambiguous Genitalia Panel includes mutation analysis of 72 genes associated with both syndromic and non-syndromic DSDs. This comprehensive panel evaluates a broad range of genetic causes of ambiguous or abnormal genitalia, including conditions in which abnormal genitalia are the primary physical finding as well as syndromic conditions that involve abnormal genitalia in addition to other congenital anomalies. -

Genetic Mechanisms of Disease in Children: a New Look
Genetic Mechanisms of Disease in Children: A New Look Laurie Demmer, MD Tufts Medical Center and the Floating Hospital for Children Boston, MA Traditionally genetic disorders have been linked to the ‘one gene-one protein-one disease’ hypothesis. However recent advances in the field of molecular biology and biotechnology have afforded us the opportunity to greatly expand our knowledge of genetics, and we now know that the mechanisms of inherited disorders are often significantly more complex, and consequently, much more intriguing, than originally thought. Classical mendelian disorders with relatively simple genetic mechanisms do exist, but turn out to be far more rare than originally thought. All patients with sickle cell disease for example, carry the same A-to-T point mutation in the sixth codon of the beta globin gene. This results in a glutamate to valine substitution which changes the shape and the function of the globin molecule in a predictable way. Similarly, all patients with achondroplasia have a single base pair substitution at nucleotide #1138 of the FGFR3 gene. On the other hand, another common inherited disorder, cystic fibrosis, is known to result from changes in a specific transmembrane receptor (CFTR), but over 1000 different disease-causing mutations have been reported in this single gene. Since most commercial labs only test for between 23-100 different mutations, interpreting CFTR mutation testing is significantly complicated by the known risk of false negative results. Many examples of complex, or non-Mendelian, inheritance are now known to exist and include disorders of trinucleotide repeats, errors in imprinting, and gene dosage effects. -
Second-Tier DNA Confirmation of Newborn Screening Results
Second-tier DNA Confirmation of Newborn Screening by Targeted Next Generation Sequencing Edwin Naylor, Ph.D. MPH Andy Bhattacharjee , Ph.D. Erik Puffenberger, Ph.D.; Kevin Strauss, MD; Holmes Morton, MD Newborn Screening & Clinical Genomics 1961 1990’s 2010-2012 2 Robert Guthrie Development of develops simple automated MS/MS Newborn Screening screening across (NBS) several disorders Current de facto standard 2 Why Newborn Genomics? • Mendelian Diseases disproportionately affect Newborns - ~3500 genetic diseases with molecular basis - >10% of NICU admissions are genetic Clinical manifestation of Genetic diseases - Current NBS tests limited to 29+ diseases CHROMOSOMAL - 2nd tier DNA testing to validate biochemical results MULTI-FACTORIAL SINGLE GENE (MENDELIAN) • Advantage of NGS based DNA testing individuals # of Affected - Find causal variants (rare/novel) in gene(s) - A ‘universal’ NGS approach avoids repeated, serial BIRTH PUBERTY ADULT single gene testing Gelehrter TD, Collins FS, Ginsburg D. Principles of - Current Sanger sequencing is expensive ($3-10K) and Medical Genetics. 2nd ed. Baltimore, MD: Williams & slow (3 months to 1 year) Wilkins; 1998:1-42 NICU- Neonatal Intensive Care Unit NBS-Newborn Screening 3 NGS-Next Generation Sequencing Why Targeted (Exome) Sequencing for now? NGS Sequencing * Genomic DNA from Causal Mutations in Affected Individuals Exons/Target Regions Fold Test Menu Cost ($) Throughput Efficiency Whole Genome (Res.) 7,666* 1 1 Exome (Res) 1,200 15 95 Neonate Panel (Clinical) <1000 150 >1140 •Majority of known disease-causing mutations in exons •Exome = protein-encoding parts of genes •Targeted NGS is Cost & Throughput Efficient *Saunders et al., (2012) Rapid Whole Genome Sequencing for Genetic Disease Diagnosis in NICUs 4 Workflow for 2nd Tier Newborn Screening Sample 2h DNA Capture 92h Raw Data 10h Analysis 1h+ Isolation & Sequencing Management & Interpretation 8 samples, 105 Hrs, <$10,000 = Real Neonatal Genomics! 5 Workflow for 2nd Tier Newborn Screening Sample •High M.Wt. -

Open Full Page
CCR PEDIATRIC ONCOLOGY SERIES CCR Pediatric Oncology Series Recommendations for Childhood Cancer Screening and Surveillance in DNA Repair Disorders Michael F. Walsh1, Vivian Y. Chang2, Wendy K. Kohlmann3, Hamish S. Scott4, Christopher Cunniff5, Franck Bourdeaut6, Jan J. Molenaar7, Christopher C. Porter8, John T. Sandlund9, Sharon E. Plon10, Lisa L. Wang10, and Sharon A. Savage11 Abstract DNA repair syndromes are heterogeneous disorders caused by around the world to discuss and develop cancer surveillance pathogenic variants in genes encoding proteins key in DNA guidelines for children with cancer-prone disorders. Herein, replication and/or the cellular response to DNA damage. The we focus on the more common of the rare DNA repair dis- majority of these syndromes are inherited in an autosomal- orders: ataxia telangiectasia, Bloom syndrome, Fanconi ane- recessive manner, but autosomal-dominant and X-linked reces- mia, dyskeratosis congenita, Nijmegen breakage syndrome, sive disorders also exist. The clinical features of patients with DNA Rothmund–Thomson syndrome, and Xeroderma pigmento- repair syndromes are highly varied and dependent on the under- sum. Dedicated syndrome registries and a combination of lying genetic cause. Notably, all patients have elevated risks of basic science and clinical research have led to important in- syndrome-associated cancers, and many of these cancers present sights into the underlying biology of these disorders. Given the in childhood. Although it is clear that the risk of cancer is rarity of these disorders, it is recommended that centralized increased, there are limited data defining the true incidence of centers of excellence be involved directly or through consulta- cancer and almost no evidence-based approaches to cancer tion in caring for patients with heritable DNA repair syn- surveillance in patients with DNA repair disorders. -

Structure, Function and Five Basic Needs of the Global Health Research System
Edinburgh Research Explorer Structure, function and five basic needs of the global health research system Citation for published version: Rudan, I & Sridhar, D 2016, 'Structure, function and five basic needs of the global health research system', Journal of Global Health, vol. 6, no. 1, pp. 010505. https://doi.org/10.7189/jogh.06.010505 Digital Object Identifier (DOI): 10.7189/jogh.06.010505 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Journal of Global Health Publisher Rights Statement: Copyright © 2016 by the Journal of Global Health. All rights reserved. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited. General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download -

Inherited Metabolic Disease
Inherited metabolic disease Dr Neil W Hopper SRH Areas for discussion • Introduction to IEMs • Presentation • Initial treatment and investigation of IEMs • Hypoglycaemia • Hyperammonaemia • Other presentations • Management of intercurrent illness • Chronic management Inherited Metabolic Diseases • Result from a block to an essential pathway in the body's metabolism. • Huge number of conditions • All rare – very rare (except for one – 1:500) • Presentation can be non-specific so index of suspicion important • Mostly AR inheritance – ask about consanguinity Incidence (W. Midlands) • Amino acid disorders (excluding phenylketonuria) — 18.7 per 100,000 • Phenylketonuria — 8.1 per 100,000 • Organic acidemias — 12.6 per 100,000 • Urea cycle diseases — 4.5 per 100,000 • Glycogen storage diseases — 6.8 per 100,000 • Lysosomal storage diseases — 19.3 per 100,000 • Peroxisomal disorders — 7.4 per 100,000 • Mitochondrial diseases — 20.3 per 100,000 Pathophysiological classification • Disorders that result in toxic accumulation – Disorders of protein metabolism (eg, amino acidopathies, organic acidopathies, urea cycle defects) – Disorders of carbohydrate intolerance – Lysosomal storage disorders • Disorders of energy production, utilization – Fatty acid oxidation defects – Disorders of carbohydrate utilization, production (ie, glycogen storage disorders, disorders of gluconeogenesis and glycogenolysis) – Mitochondrial disorders – Peroxisomal disorders IMD presentations • ? IMD presentations • Screening – MCAD, PKU • Progressive unexplained neonatal -

The Genetic Basis for Skeletal Diseases
insight review articles The genetic basis for skeletal diseases Elazar Zelzer & Bjorn R. Olsen Harvard Medical School, Department of Cell Biology, 240 Longwood Avenue, Boston, Massachusetts 02115, USA (e-mail: [email protected]) We walk, run, work and play, paying little attention to our bones, their joints and their muscle connections, because the system works. Evolution has refined robust genetic mechanisms for skeletal development and growth that are able to direct the formation of a complex, yet wonderfully adaptable organ system. How is it done? Recent studies of rare genetic diseases have identified many of the critical transcription factors and signalling pathways specifying the normal development of bones, confirming the wisdom of William Harvey when he said: “nature is nowhere accustomed more openly to display her secret mysteries than in cases where she shows traces of her workings apart from the beaten path”. enetic studies of diseases that affect skeletal differentiation to cartilage cells (chondrocytes) or bone cells development and growth are providing (osteoblasts) within the condensations. Subsequent growth invaluable insights into the roles not only of during the organogenesis phase generates cartilage models individual genes, but also of entire (anlagen) of future bones (as in limb bones) or membranous developmental pathways. Different mutations bones (as in the cranial vault) (Fig. 1). The cartilage anlagen Gin the same gene may result in a range of abnormalities, are replaced by bone and marrow in a process called endo- and disease ‘families’ are frequently caused by mutations in chondral ossification. Finally, a process of growth and components of the same pathway. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Congenital Disorders of Glycosylation from a Neurological Perspective
brain sciences Review Congenital Disorders of Glycosylation from a Neurological Perspective Justyna Paprocka 1,* , Aleksandra Jezela-Stanek 2 , Anna Tylki-Szyma´nska 3 and Stephanie Grunewald 4 1 Department of Pediatric Neurology, Faculty of Medical Science in Katowice, Medical University of Silesia, 40-752 Katowice, Poland 2 Department of Genetics and Clinical Immunology, National Institute of Tuberculosis and Lung Diseases, 01-138 Warsaw, Poland; [email protected] 3 Department of Pediatrics, Nutrition and Metabolic Diseases, The Children’s Memorial Health Institute, W 04-730 Warsaw, Poland; [email protected] 4 NIHR Biomedical Research Center (BRC), Metabolic Unit, Great Ormond Street Hospital and Institute of Child Health, University College London, London SE1 9RT, UK; [email protected] * Correspondence: [email protected]; Tel.: +48-606-415-888 Abstract: Most plasma proteins, cell membrane proteins and other proteins are glycoproteins with sugar chains attached to the polypeptide-glycans. Glycosylation is the main element of the post- translational transformation of most human proteins. Since glycosylation processes are necessary for many different biological processes, patients present a diverse spectrum of phenotypes and severity of symptoms. The most frequently observed neurological symptoms in congenital disorders of glycosylation (CDG) are: epilepsy, intellectual disability, myopathies, neuropathies and stroke-like episodes. Epilepsy is seen in many CDG subtypes and particularly present in the case of mutations -

NICU Gene List Generator.Xlsx
Neonatal Crisis Sequencing Panel Gene List Genes: A2ML1 - B3GLCT A2ML1 ADAMTS9 ALG1 ARHGEF15 AAAS ADAMTSL2 ALG11 ARHGEF9 AARS1 ADAR ALG12 ARID1A AARS2 ADARB1 ALG13 ARID1B ABAT ADCY6 ALG14 ARID2 ABCA12 ADD3 ALG2 ARL13B ABCA3 ADGRG1 ALG3 ARL6 ABCA4 ADGRV1 ALG6 ARMC9 ABCB11 ADK ALG8 ARPC1B ABCB4 ADNP ALG9 ARSA ABCC6 ADPRS ALK ARSL ABCC8 ADSL ALMS1 ARX ABCC9 AEBP1 ALOX12B ASAH1 ABCD1 AFF3 ALOXE3 ASCC1 ABCD3 AFF4 ALPK3 ASH1L ABCD4 AFG3L2 ALPL ASL ABHD5 AGA ALS2 ASNS ACAD8 AGK ALX3 ASPA ACAD9 AGL ALX4 ASPM ACADM AGPS AMELX ASS1 ACADS AGRN AMER1 ASXL1 ACADSB AGT AMH ASXL3 ACADVL AGTPBP1 AMHR2 ATAD1 ACAN AGTR1 AMN ATL1 ACAT1 AGXT AMPD2 ATM ACE AHCY AMT ATP1A1 ACO2 AHDC1 ANK1 ATP1A2 ACOX1 AHI1 ANK2 ATP1A3 ACP5 AIFM1 ANKH ATP2A1 ACSF3 AIMP1 ANKLE2 ATP5F1A ACTA1 AIMP2 ANKRD11 ATP5F1D ACTA2 AIRE ANKRD26 ATP5F1E ACTB AKAP9 ANTXR2 ATP6V0A2 ACTC1 AKR1D1 AP1S2 ATP6V1B1 ACTG1 AKT2 AP2S1 ATP7A ACTG2 AKT3 AP3B1 ATP8A2 ACTL6B ALAS2 AP3B2 ATP8B1 ACTN1 ALB AP4B1 ATPAF2 ACTN2 ALDH18A1 AP4M1 ATR ACTN4 ALDH1A3 AP4S1 ATRX ACVR1 ALDH3A2 APC AUH ACVRL1 ALDH4A1 APTX AVPR2 ACY1 ALDH5A1 AR B3GALNT2 ADA ALDH6A1 ARFGEF2 B3GALT6 ADAMTS13 ALDH7A1 ARG1 B3GAT3 ADAMTS2 ALDOB ARHGAP31 B3GLCT Updated: 03/15/2021; v.3.6 1 Neonatal Crisis Sequencing Panel Gene List Genes: B4GALT1 - COL11A2 B4GALT1 C1QBP CD3G CHKB B4GALT7 C3 CD40LG CHMP1A B4GAT1 CA2 CD59 CHRNA1 B9D1 CA5A CD70 CHRNB1 B9D2 CACNA1A CD96 CHRND BAAT CACNA1C CDAN1 CHRNE BBIP1 CACNA1D CDC42 CHRNG BBS1 CACNA1E CDH1 CHST14 BBS10 CACNA1F CDH2 CHST3 BBS12 CACNA1G CDK10 CHUK BBS2 CACNA2D2 CDK13 CILK1 BBS4 CACNB2 CDK5RAP2